Thoughts from the President
June 27, 2025 - Transitions in Leadership
Dear Danforth Center Community,
Becky Bart’s departure later this summer means we’ll be saying farewell to an exceptional scientist, mentor, and valued contributor at the Danforth Center. As a long-time research collaborator with Becky and a friend, this is a tough one for me. Her departure also means we will need a new Vice President for Research (VPR), a role Becky has filled with skill, vision, patience, energy and compassion for the past 18 months. I cannot thank Becky enough for serving the Center as a leadership partner with me. But as difficult as departures may be, they are also opportunities for others to step up and contribute in new ways.
Given the other leadership changes happening in 2025, we set out to identify and appoint an interim VPR from within the senior faculty ranks through a nomination process. We considered the possibility that the job could be done by interim co-VPRs in a shared appointment. I was highly encouraged that eight individuals stepped forward, a positive sign that many faculty are seeking ways to serve the broader Danforth Center community. An advisory group composed of Tessa Burch-Smith, Anna Dibble, Jim Umen and Becky provided insight and wisdom during the selection process.
I am thrilled to announce that Allison Miller and Keith Slotkin have agreed to serve as interim co-VPRs. Starting next week, they will serve as Leadership Team members, oversee discretionary research funding programs, lead faculty promotion and retention processes, oversee core facility directors, and more. Both are scientific leaders at the Center and in the broader plant science community, and each has unique ideas and strengths to help lead our scientific enterprise forward. They also appreciate the importance to stability in the near and intermediate future as we navigate many changes.
They assume new roles despite already having numerous ongoing responsibilities. I sincerely thank Allison and Keith for taking on the challenge and serving in this capacity. I’m excited to work closely with them and to see what they achieve.
Jim Carrington,
President and Chief Executive Officer
Previous Weekly Messages from Jim Carrington
Dear Danforth Center Community,
There are few things are more satisfying that being recognized, awarded, and rewarded by your professional peers. I mentioned a few awards in the March 14 weekly message, so let’s catch up and highlight Danforth Center community members who have been honored recently or that I missed earlier.
2025 CSTM Professional Development Awards. The Committee for Scientific Mentoring and Training coordinates awards to non-faculty scientists at the Center to offset costs of professional development (e.g. travel to workshops and conferences.) Congratulations to Mazen Alazem, Spencer Arnesen, Keith Duncan, Myia Elliott, Patricia Gallardo, Kerri Gilbert, Kirsten Hein, Chloe Hendrikse, Ray Kannenberg, Ketra Oketcho, Vidhyavathi Raman, Parbati Thapa, and Elizabeth Vanhoecke on awards announced last month. Also, congratulations Anna Casto and Andrea Zanini for being honored as this year’s IWISH (International Women in Science Hub) CSTM Travel Award recipients.
Graduate Fellowship Awards. The Danforth Center now recognizes two graduate students annually with competitive fellowships. The 2025 Danforth Plant Science Fellowship was awarded to Washington U. Ph.D. student Sam Nuzzi (Burch-Smith lab), who is working to understand mechanisms of intercellular trafficking in plants. The inaugural William T. Kemper Plant Science Fellowship was awarded to Washington U. Ph.D. student Olivia Gomez (Umen lab) to support her work on phosphate uptake and accumulation by plant cells. Congratulations to Sam and Olivia!
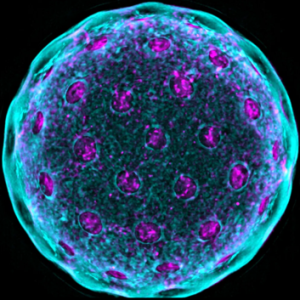
Outstanding Image Award. The BioImaging North America society recently recognized Anastasiya Klebanovychfor producing one of the three top images of the year. A highly talented scientist and manager in the Center’s Advanced Bioimaging Lab, Anastasiya used the Zeiss Elyra 7 microscope and 3-D reconstruction techniques to create a stunning image of a quinoa pollen grain (see below). Well done, Anastasiya!
ASPB Awards. The American Society of Plant Biologists named Kevin Cox (Assistant Member and PI) as the 2025 Eric E. Conn Young Investigator Award recipient. The award recognizes an outstanding plant scientist who has also demonstrated excellence in outreach, public service, mentoring, or teaching early in their faculty career. And Ivan Baxter (Member and PI) was recognized as a ASPB Fellow for direct service to the Society and distinguished, long-term contributions to plant biology. Congratulations to Kevin and Ivan!
We are a community of creative, high-achieving scientists and professionals, and it thrills me to see this kind of recognition.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
When visitors come to the Danforth Center, I often point to our prairie landscape to illustrate an important point: those plants thrive, year after year, despite no irrigation water, no pesticides, and no fertilizers. If we can understand how those native prairie plants have adapted to the stresses of temperature extremes, drought, pests and diseases, we can apply that knowledge to improve the resilience of agricultural crops. With five review articles, the June 12, 2025 issue of Science (pp. 1146-1173) focuses on the challenge of understanding mechanisms underlying heat stress adaptation in plants, and the needs to quickly apply that knowledge to safeguard crop productivity in a warming world.
The articles cover a lot of ground, but I draw your attention to three themes concerning heat stress on plants:
It’s a difficult problem to understand. Much work has focused on the molecular details of stress responses in model plants, like Arabidopsis, and in a few major crops. But what we’ve learned does not always scale to understand how plants in agricultural and complex ecosystems cope with heat stress. Also, while it is clear that can plants evolve and adapt in heat-stressed environments over long periods of time, different plant species do these in very different ways. That means improving heat stress tolerance in different crops will likely require diverse approaches.
Heat stress affects the plant microbiome. Plant microbiomes are greatly affected by heat stress. In one study, the abundance of beneficial mycorrhizal fungi, which help plants take up nutrients from soil, was decreased by 59% at elevated temperatures. In contrast, several types of disease-causing bacteria and fungi grow better and become bigger problems at high temperatures. In other words, rising temperatures mean that crop-microbe interactions become less suited for productive agriculture.
Heat stress affects photosynthesis. As an insightful person said recently, “Plants feed the world and the sun feeds the plants.” Photosynthesis, the mechanism whereby green plants capture energy from the sun, is inhibited as temperatures rise above 40C. Research points the way towards developing plants with more resilient photosynthetic mechanisms at high temperatures. Safeguarding photosynthesis through new crop traits may be necessary in the near future.
With the goal of mitigating the worsening effects of rising temperatures, the Danforth Center has considerable ongoing research in these areas. It is important work that’s needed for a food-secure future.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
In 2002, our team at Oregon State University (OSU) published a series of early papers on small RNAs (siRNAs and microRNAs) in plants, and we attracted a lot of colleagues and collaborators who wanted to learn our methods. One such colleague was a young, German Ph.D. student from a lab across campus. He made several working visits and gained what he needed from Kristin Kasschau, an exceptional senior scientist who was happy to help. I met the student, but honestly, it faded from memory shortly thereafter.
Fast-forward to the Danforth Center atrium yesterday and the 10-year anniversary celebration for the KWS Gateway Research Center, located at BRDG Park. KWS is a major, global company that develops and provides elite seed for corn, sugar beet, and many cereal and vegetable crops. Now with 50 local employees focused on advanced trait development on our campus, KWS has been a vital member of the AgTech ecosystem in 39North and St. Louis since 2015. My role at the event was to get it going with a welcome, say a few words about our partnership, and give thanks to KWS team members and leadership, including their CEO, Felix Büchting. Felix is the great-great-great-great grandson of the founder of KWS, and is continuing the family tradition of leading the company. I did not recall ever meeting Felix previously, so I arrived early to introduce myself and have a brief chat.
“Hi, Felix, it’s good to meet you,” I said while shaking hands. “Yes, it’s good to meet you again,” Felix replied. Now, the instant he said the part about “again,” two urgent thoughts started competing for brain time: Where have we met before, and how old am I getting? With a big smile on his face, Felix explained that he spent time in our lab at OSU, learning how to analyze small RNAs while he was a graduate student in the early 2000s. Felix then said that the Germans have a saying: Man sieht sich immer zweimal im Leben. You always meet twice in a lifetime.
Yesterday’s celebration with KWS was a reminder of the relevance of our mission, and the importance of our neighbors. It also got me thinking about how much better we could all be if we started every individual encounter with the assumption that we would meet that person later.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Every year in late May, I meet with the summer research interns at the beginning their 10-week Danforth Center program. The undergraduate intern program has been running for over 20 years, and was recently renamed Field and Laboratory Learning in Plant Science (FLLiPS). We are fortunate to host 17 ambitious students from colleges and universities in 10 states around the U.S. Several students originate from the St. Louis area. I asked the interns about their career interests in plant science, and how they envision using it to address big challenges in the world. Their impressive responses affirm that they chose the right place to spend their summer.
They are interested in genetics and genome editing/engineering. Several interns, like Anna Meirink (Loyola University), see great benefits in using genetics to improve crops. “It’s really fascinating that you can introduce mutations in plants to make them resistant to viruses, for example,” said Anna. I couldn’t agree more.
They want to get advanced degrees. The vast majority of interns said they expect to attend graduate school. They’ve been bitten by the research bug and they want to build careers armed with a Ph.D. Several interns have experience already doing research in labs, while others are just getting started.
They are interested in learning and using technology. Several interns will spend their summer working with data science or imaging technologies like x-ray tomography. ZhakeYa Hawkins (Kentucky State University) said, “I’m interested in anything using computers and data science to understand plants.” ZhakeYa knows what’s possible, because she has already done work at the Danforth Center as a high school student through the JJK-FAN program in her hometown of East St. Louis.
They want to contribute to more sustainable agriculture. They recognize the critical importance of applying science to lower the environmental footprint of agriculture. Several interns mentioned their interest in helping to develop more water use-efficient and drought-tolerant crops.
They see plant science as a positive contributor to regional development. Through new innovations, good jobs and economic develop, the interns recognized that plant science can enhance a region. We’ve certainly seen that here in the St. Louis area.
The 2025 FLLiPS program and students are supported by funding through the NSF-Research Experiences for Undergraduates, the Saigh Foundation, and several grants to Danforth Center PIs. I thank all who are serving as research mentors for interns, as well as Tessa Burch-Smith, Kirk Czymmek, Marisa Yoder, Becca Bindbeutel and Genetta Robinson for running this incredible program.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
What a difference a day makes. What started last Thursday as a celebration of amazing talent and creativity at Big Ideas turned into a nightmare 20 hours when a deadly storm and tornado struck St. Louis. So many Danforth Center community members and their families suffered damage to, or loss of, their homes, cars, property and electrical power along the tornado’s path. Some were spared a direct hit but sustained massive amounts of damage in their neighborhoods.
I want to recognize and thank all of the Danforth Center community who joined with friends, neighbors, colleagues and others across the region to offer relief to those in need. Some donated time and hard work to clear debris from neighborhoods. Some offered their homes to displaced friends and neighbors, or to those without power. Some donated money, food, clothing and supplies to food banks and relief centers. Clara Lebow and Kirsten Hein organized collections that filled at least 2.5 large bins at the Danforth Center. Many of you made contributions to the Danforth Center Cares fund, which provides support to those in need within the Center community. Some Danforth Center labs and departments collected donations specifically for affected team members.
We’re a generous community that shows how much we care about our colleagues in numerous ways, from showing up to celebrate Big Ideas teams to helping those who suffered losses from a random, destructive tornado. That contributes to our culture and resilience as a Center community, in good times and bad times.
Speaking of resilience, at least five Danforth Center community members who suffered significant damage to their homes joined 15 others on a Center team as part of the 2025 Biz Dash 5K last evening in downtown St. Louis. Anna Casto, one of the runners, a Big Ideas speaker and a resident with damage along the tornado path, said to me just before the race, “Big Ideas seems so long ago with everything that’s happened since.” She then led the Danforth Center team, along with Noah Fahlgren, Becky Bart and Jorge Gutierrez, with team-best times to win the Foundations and Non-profits category, and place 8th overall out of 242 teams.
Jim Carrington,
President and Chief Executive Officer
P.S. Congratulations to Team Sun Solutions for winning the audience vote for favorite presentation!
Dear Danforth Center Community,
I admire those who, despite their reservations or uncertainties, dive in head-first to solve a problem or do something different. My admiration meter hit maximum level last night at Big Ideas where Vanessica Jawahir, Salma Adam, and Marcus Griffiths (Team Sun Solutions); Keely Brown, Josh Sumner and Anna Casto (Team Bioqumulate); and Sam Nuzzi, Ally Angermeier and Anastasiya Klebanovych (Team Pore Decisions) gave three amazing presentations. For those unfamiliar with Big Ideas, each team presented an inspiring 13-minute talk about an impactful idea that radiates from their Danforth Center research. The talks were accessible, entertaining and meaningful to a public audience. The teams also answered three or four questions from a lively panel composed of Penny Pennington, Tom Hillman and Arnold Donald, who had no advance knowledge of the topics or content. Katie Murphy served as a magnificent emcee for the event.
Over 10-weeks of preparation, the teams honed ideas, crafted content, and developed, revised, and rehearsed presentations; most team members were pushed outside of the comfort zones. But in front of 450 people onsite, each team was brilliant! They may have even surprised themselves. As an interested observer of the process they experienced, I have a few thoughts about why they did so well.
They started with why. Not only did each team start with the big problem they were addressing – improving crop productivity, remediating toxic waste sites, or preventing plant diseases – they led with personal stories and experiences that brought relevance to everyone. They immediately gave reasons to care and built strong connections with the audience.
They worked as teams. Each team member leaned into their own strengths and experience. They did not simply sing together, they harmonized. And despite the fact that Big Ideas was a competition, each team also helped the others by offering countless suggestions and feedback.
They used all of their tools. On stage, the teams developed rapport and figured out ways to connect with each other through physical responses, verbal reactions, and humor. They used humor creatively and effectively to make their case and engage the audience. And as one viewer said, “I think what was most important was how likeable and disarming they all were.” Great presentations involve far more than the words.
They worked hard. Any comments about how hard they worked, on top of their daily responsibilities, would be an under-statement.
I thank each Big Ideas team member for trusting yourselves, your teammates and your ideas, and for diving in head first!
Jim Carrington,
President and Chief Executive Officer
P.S. Congratulations to Team Sun Solutions for winning the audience vote for favorite presentation!
Dear Danforth Center Community,
A few weeks ago, we convened our annual “Faculty Retreat” at the Jones Visitor and Education Center at Forest Park. This one-day event serves to explore, brainstorm, and raise concerns about issues, opportunities or changes that affect the Danforth Center faculty and their team members. To encourage follow-on discussions within teams, here’s what we discussed.
Internal funding opportunities. Internal research funding opportunities are becoming more and more important. We discussed how to access the Proof-of-Concept fund, Institutional Equipment Fund, graduate student fellowships, and other programs. Last year’s Retreat included a poll to assess the most needed kinds of internal funds, with the top results being 1) startup fund replenishment, and 2) graduate student support. Decisions were made soon thereafter to reprioritize some reserves to re-fund startup accounts and to elevate new fundraising for additional fellowships.
Developments within SINC Center. The Subterranean Influences on Nitrogen and Carbon (SINC) Center seeks to develop technologies to reduce the use and impact of synthetic nitrogen fertilizer in agriculture. The team has developed a number of resources, like the Elementar element analyzer and long-term cover crop study site at the Field Research Site. We heard how the SINC team is looking to increase and incentivize engagement with additional Danforth Center teams.
Spinning out companies through the Startup Initiative. Three companies have been spun out through Danforth Technology Company (DTC) since 2021. We heard the unique stories from company co-founders for how the idea for each one originated, how Center resources were used to develop the underlying science, and how the co-founders worked with DTC to create the companies.
Grant proposals, funding, mitigation strategies, and policies. Much time was spent discussing and gathering ideas for how to improve our proposal submission processes, including ways to reduce the frequency of missed internal deadlines. We discussed ways in which we plan to mitigate federal grant reductions, including those presented in prior weekly messages. Given the highly collaborative nature of the grant preparation and administration processes, we need ideas and cooperation from across the Center to navigate forward.
Leadership transition. The upcoming transition of Danforth Center President is forefront on many minds. At the Retreat, I appreciated the ideas and focus on ways in which the faculty can contribute to a smooth transition.
I want to underscore the value in hearing diverse viewpoints through activities like the Faculty Retreat. Not only does this elevate understanding, but it helps improve decision-making. I thank all of the Retreat participants, including PIs, directors, CSTM co-chairs, the leadership team, and the organizers.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
As in most weeks during these chaotic times, there were both good reasons to celebrate the Danforth Center community and our work, as well as opportunities to display resilience and exercise patience. I want to update the community on a few items across both categories.
Annual Engagement Survey – We received results this week and were thrilled to see over 71% of employed community members complete the 2025 survey. That was significantly higher than the 52% participation from last year. Our “Overall Workplace Experience Score” increased to 78%, up 5 percentage points from last year and 6 points above the benchmark for organizations similar to ours. Not only were scores up across most categories, we received 649 written comments. I sincerely thank everyone who took the time and effort to participate in this year’s survey. The team from People and Culture will be in touch next week with much more information about Survey results and ways in which you can provide more feedback and input.
Concerns about the National Science Foundation – The NSF is one of our top three funding agencies. For the past few months, we’ve been experiencing delays in both activation of new NSF awards and implementation of incremental funding (e.g. funding for the next year of a multi-year project,) as was also described this morning in a news article posted at Nature. For example, incremental funding for the 2025 Summer Research Experiences for Undergraduate (REU) program has been delayed with no timeline to activation. And as was expected, NSF today announced an order to cap indirect cost rates at 15% of direct costs, though we also expect that order to be held up in court. We are monitoring the situation at NSF carefully and communicating our serious concerns to elected officials.
Encouraging support from the Saigh Foundation. I communicated recently about redirecting some of the Future Forward fundraising effort towards mitigating potential loss of federal grant funds, and how private donors and foundations are more important than ever to the Center. The redirection has already begun, and we’ve been encouraged by responses from several donors. The Saigh Foundation, for example, recently provided funding to support five REU students this summer. When combined with other funding sources, this will enable the program to train 17 students this year, even without incremental NSF support. The Foundation has also made a multi-year commitment that enables the Danforth Center to support REU or other training activities well into the future. Many thanks to Lee’at Bachar in the Development team for above-and-beyond work with the Saigh Foundation.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Early last year, we formed an ad hoc Danforth Center Art Committee to identify new pieces to display on the second level, adjacent to the main elevator. The committee, composed of Kerri Gilbert, Sarah Jennings, Anna Dibble and Todd Hornburg, pondered ideas that would be authentic representations of the Danforth Center and our values, be beautiful and creative, and be inspiring to all who view the art. The group quickly latched on to the notion of art that incorporates plant material, and soon thereafter, the committee was scrolling through websites and saying, “Ooh, look what you can do with a moss wall!”

That led the group to Lynne Young, a St. Louis moss wall artist who also works at Growing Green, the company that takes care of the Center’s indoor potted plants. Lynne describes her work as watercoloring with moss. The committee asked Lynne, “Could you create a representation of a Missouri tall grass prairie, like the one at the Danforth Center?” To say Lynne was immediately comfortable with the idea, or that she understood what we had in mind, might not be so accurate. But over the course of nearly one year, 8-10 meetings with members of the committee, and much experimentation, Lynne figured out how to turn three types moss, different shades of barba grass, and vegetative and floral parts of yarrow, caspia, gyposphilla, glixia and sea lavender plants into two spectacular images of a prairie in bloom. These were the most elaborate and unusual pieces Lynne has ever created.
Anna loves that the art “reflects the beauty of our prairie and the spirit of our community. This unique installation was thoughtfully designed just for us, making it a great example of innovation.” When asked what was most fun about this project, Kerri said, “All of the ideas that we brainstormed as possible new artwork, and seeing the process of going from concept to this beautiful art.” Lynne said, “Seeing people’s faces light up and saying ‘wow’ after we installed it was the best part.”
I hope everyone enjoys viewing and showing the new artwork, and sees a little bit of themselves in what it represents. I sincerely thank Kerri, Sarah, Anna, Todd and Lynn for giving us something special.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
You may have seen that we publicly announced the Future Forward campaign last week. Future Forward seeks to raise at least $150M from philanthropic sources to support the Danforth Center’s work, strengthen our institution and grow our impact. The campaign was designed to support priorities within the current strategic plan, so the fundraising priorities of Future Forward will be familiar to those in the Danforth Center community. In short, we seek to raise funds to support 1) delivery of improved crops for smallholder farmers; 2) research, facilities and initiatives to enable more sustainable agriculture; 3) innovation leading to new companies and a trained workforce in the region; and 4) core strength and robustness of the Center’s people, facilities and programs.
Future Forward has been active during a pre-announced, quiet phase for several years, so we are well-along towards achieving our goal. In fact, you have seen much impact from campaign fundraising already, including the Field Research Site acquisition, development and implementation. We funded the Startup Initiative to create new companies based on Danforth Center science and technology; three companies have been spun out as a result. The SINC Center was created to develop new ways to lower the use of synthetic nitrogen fertilizer in crop agriculture, and several core facilities have implemented new equipment and technologies. Our endowment has grown significantly through campaign gifts, which support trainees, graduate students and all functions across the Center. Everyone at the Danforth Center has already benefited from Future Forward.
With the uncertainties and reductions at federal agencies, funds raised through Future Forward will be critical to mitigate effects of potential grant funding declines. Campaign fundraising has enabled creation of reserves to ensure that key priorities remain funded, startup accounts are replenished, and bridge funding is available. In fact, we have recently adjusted some campaign priorities to account for these growing needs.
The Future Forward campaign is led by an incredible Development team, but its success also depends on the rest of us. Everyone can help by participating at events and meeting donors, giving tours, sharing your stories, or contributing financially. The future of the Danforth Center will be better as a result, and for that I am sincerely thankful.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
In a public letter released by nearly 2000 members of the National Academies of Science, Engineering and Medicine earlier this week, alarms were sounded in response to the dismantling of critical national science infrastructure and research funding agencies. The letter-signers make the case that these actions will have long-lasting negative consequences on health, safety, economic competitiveness, and standards of living. Few scientists, engineers and physicians would disagree. But while I strongly support the content of the letter and the act of speaking up, I questions if this the most effective group to be making the public case? Probably not.
The decline of public trust in science, and in science-based institutions like the NIH and universities, creates a big problem for scientists serving as the messengers. First, we’re associated with the institutions in which the public increasingly perceives as insular and elitist. And second, we’re accused of distorted self-interest, like wanting to maintain our research grants. Though I believe the loss of national scientific infrastructure could be just as damaging as loss of highway, communication or airport infrastructure, it’s hard to deny that I’m not highly bothered by the prospect of losing grant funding.
More than any time in the past 80 years, we need non-scientists delivering the message in support of science. Every time we host public events at the Center, like the one we held last evening, they’re attended by interested and motivated non-scientists like financial professionals, business owners, and family farmers. Everyone I spoke with left the event more excited about the need for science to improve lives. These are the kinds of individuals who can deliver a potent message about the necessity for science and science-based decision-making at the local, state and national levels. Our challenge is scaling up this kind of engagement, changing the notion of what it means to support science, and inspiring non-scientists to amplify the message to peers and leaders.
The scientific community needs to continue speaking out, though in more human and relatable ways. We also need a far better coalition with non-scientists, because they may have the most compelling voice. Here’s to the non-scientists!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I recently ran into Kong Wong, Postdoctoral Fellow in the Topp Lab, and we chatted about his upcoming move to Virginia Tech University. Kong will be an Assistant Professor in the Department of Biological Systems Engineering where he will initiate research on biochemical engineering of plant-microbe interactions to advance sustainable agriculture. Soon after the conversation started, Kong asked, “Do you have any advice for me?” I find there are three types of people who ask this question: those who seek meaningful advice and perspective; those who seek magic formulas and pixie dust; and those who seek validation of their own views. Kong clearly resides with the first group, so after a brief but tense flashback through 23 years as a university professor, here’s what I offered.
Know what you want to achieve. Thinking through and knowing what you want to accomplish, and why, goes a long way! Clearly articulating what success looks like in your research program will inspire students and trainees, help reviewers of your grant proposals, and help you set ambitious but reachable goals. I suggested to Kong that getting tenure at a university is hardly worth worrying about. Getting tenure will be a byproduct of high achievement, so if you have to worry, worry about achieving.
Stay focused on what’s important. Universities are great places, though they are loaded with distractions. Research, teaching and training are among the many important things Kong will need to do. Constantly asking and honestly answering the question, “Is the activity I’m doing really important for what I want or need to achieve?” is a pretty good way to help minimize distractions. Never confuse activity for achievement, as John Wooden once said.
Use your time well. What else could be more obvious, yet so difficult to do? Good time management is simply the process of exercising self-discipline when you have too many options for how to spend your time. Allocating enough time to do a good job on the important things, and then actually using that time wisely, is difficult because we overbook ourselves, we check devises too often, we get bored, we have emergencies that arise, and on and on. Short of having a personal concierge who enforces time management, there are many tools and techniques that help. I have a “No Friday meetings” policy that, among other benefits, ensures that the Weekly Message gets written before the week ends.
Good luck, Kong. I’m excited for you!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Let’s take a few moments this week to celebrate a key milestone, congratulate some recent award winners, and thank a group of Danforth Center community members who go above and beyond.
Major milestone. Kevin Cox reach a significant career milestone with publication of the first original research article from his new lab. The collaborative paper integrates work from the Cox, Nusinow, Czymmek and former Meyers labs, with contributions from team members Sarah Pardi, Lily O’Connor, Anastasiya Klebanovych and David Huss. The first paper is a big deal because it establishes Kevin as an independent force in plant biology, and his lab as a destination for scientists and trainees who want to make discoveries and build their careers. The Plant Journal paper, titled “ExPOSE: a comprehensive toolkit to perform expansion microscopy in plant protoplast systems,” focuses on development of new methods that enable better localization of macromolecules in plant cells. Congratulations to Kevin and team!
Recent awards. Several Danforth Center faculty were announced recently as award winners by various scientific organizations. Katie Murphy was honored with the 2025 Early Career Award from the North American Plant Phenotyping Network. The Academy of Science of St. Louis is honoring both Allison Miller and Bing Yang at their upcoming annual awards celebration, with Allison receiving the George Engelmann Interdisciplinary Award for Collaborative Science Achievement. Bing will take home the James B. Eads Award for Excellence in Engineering or Technology. And Erin Sparks, incoming joint faculty member between Mizzou and the Danforth Center, received the M. Rhoades Early-Career Award at the annual Maize Genetics Meeting last week. These awards recognize outstanding achievements by exceptional Danforth Center colleagues. I’ll recognize non-faculty award winners separately at a later date.
Going above and beyond. On this Grant Professionals Day, let’s celebrate all those who do the hard work that enables the Center to submit proposals, manage awarded grants and contracts, and ensure that we comply with the rules. For sponsored research grants, special thanks go to the outstanding Grant Specialist team of Denise Cunningham, Alex Durdello (promoted to Sr. Grant Specialist), Dana Lorenz, Elizabeth Martinez, Julie Witzel and Natalie Piper. Wait, what about Missy Rung-Blue? Just moments ago, Missy was announced as the new Manager of Grant & System Administration, a promotion after many years of terrific work. In addition, the talented gift officers and team members in Development do the relationship-building, cultivation and heavy lifting for proposals to philanthropic organizations and individuals, and the professionals in Finance do great work to manage and oversee grant funds at pre- and post-award stages. Many thanks to all of our grant professionals!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
A committed group of Danforth Center community members are attending the Stand Up for Science rally in Jefferson City this afternoon. Stand Up for Science events will be happening today in capital cities around the U.S. to show grassroots support for scientific research, science education, and preservation of science funding. The events show that we care, and that we are many! The importance of scientists and science supporters being seen and heard by elected officials and the public has never been higher. This platform and mass show of support focuses attention on the vital importance and impact of science, and I thank each participant for your presence and voice today.
I appreciate the diverse ways in which individuals connected to the Center are standing up for science in public through their work, volunteer activities, and passions. Consider Marilyn Bush (Bank of America) and Benjamin Akande (Stifel Financial), key executives at financial institutions here in St. Louis and the newest Directors elected to the Danforth Center Board. By serving on our Board, they are standing up for science by helping us deliver on our mission. They have stood with us through the years by helping to raise funds from companies and donors, and volunteering at our public events like Big Ideas and Seeds of Change. Marilyn has even nominated scientists for awards in the business community, enabling the importance of work like ours to be highlighted and appreciated by new audiences. Thank you, Marilyn and Benjamin, for many more years of standing up for science with the Danforth Center!
How about Kristina DeYoung, communications specialist in the Public Relations team? Kristina is a member of The Opera Bell Band, a theatrically inclined group that has at least 30 minutes of music and performance content about corn! Who knew? In her talented way, Kristina is using the stage and microphone to bring a little more awareness about plants to audiences that are not expecting it. Thank you, Kristina, for standing up and performing in a way that honors science. By the way, if you’re attending the 67th Annual Maize Genetics Meeting being held this week at Union Station in St. Louis, The Opera Bell Band is performing their salute to corn this evening.
We all have platforms and megaphones, big or small, to stand up for science. Whatever yours are, we need to use them more than ever!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
The outlook for federal grant funding to support research and training at the Danforth Center is weakening. Our funding model currently depends on competitive grants from federal agencies, grants from private foundations and companies, and grants or gifts from philanthropic donors. Even though the extent of potential losses of federal grants is not at all clear, I want to explain a little more about how we could be impacted and the ways in which we could potentially adjust.
It is important to understand that federal grant dollars for projects fund two general types of costs. The first is direct costs, which simply cover the expenses for personnel, supplies, use of greenhouse or field space, and other needs to get a specific project done. The second is indirect costs (IDCs), which cover the fraction of facilities and administrative expenses required to do the work, but which are not specific to that project. Facilities costs include things like maintenance of labs, some shared equipment, and electricity, while administrative costs include those for finance and compliance support, human resource management, and other necessary services. (Note: IDCs do not include costs that are unrelated to research, like those for the Development team.) The amount of IDC funding in federal grants, which for us is 65% of direct costs, is based on a rate negotiated with, and approved by, the federal government based on the actual costs we incur. In contrast to how they are erroneously described by some detractors, IDC funds cover real costs that enable the Danforth Center to function as a productive plant science research organization.
Significantly less federal grant funding through fewer grants and/or a mandated lower IDC rate puts important research capacity and critical administrative functions at risk. That begs the question: What would we do if this happens? Both long- and short-term adjustments would likely be necessary. We could seek more research funding from non-federal sources; nearly 50% of research funding already comes from such sources. We could seek a higher percentage of overall funding from philanthropic donors. We could redirect funds that are currently in hand for capital or building projects to cover more acute needs. We maintain an operating reserve that could potentially be tapped in the event of an operating shortfall or an emergency. And, we could lower costs by making some difficult choices.
This message is not intended to convey gloom and doom, but rather, to build early awareness and inform about the tools we have to mitigate or reduce the impact of losses in federal grant funding. Changes may be needed, but we’re in good shape to make them.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Someone asked me recently, “Are you worried that the Danforth Center is going to change with a new President?” This question was asked by a highly committed Danforth Center community member who has contributed more than their fair share to what makes the Center special. It was asked to understand how I was feeling about stepping down, and also to gauge if I was concerned about losing some things that we’ve worked hard to achieve over the past 14 years. Here’s my answer: No, I am not. I would be worried if the Danforth Center does not change!
First of all, the state of scientific research changes, and rapidly! New leadership provides the opportunity to add fresh perspective, stimulate new ideas, and make new investments that enable meaningful scientific progress at the leading edge to continue. Second, technology changes at an accelerating pace, and technology is a prime enabler of scientific discovery. New leadership can enable the push and the space to rethink, and invest in, the technological changes needed for both scientific and administrative parts of the Center. These kinds of changes happened over the past 14 years and helped get us to today, but those changes are not sufficient to get us to tomorrow.
Third, the speed with which our work yields real impact in the world will need to accelerate. Danforth Center research that enables meaningful impact can require decades, as well as patient investors. We will need to accelerate movement toward proof of concept and prototypes, and to develop faster ways to take those innovations to scale, whether in underdeveloped regions of the world or those with advanced markets.
And fourth, how we fund the work of the Center may need to change. There’s a real possibility that funding we receive through competitive grants from the federal government will decline significantly. This would mean that a greater proportion of funding for scientific projects may need to come from philanthropic donors, foundations and other sources. Our culture of philanthropy, which we value greatly at the Center, must continue to strengthen.
I cannot imagine a better leader than Giles Oldroyd, Danforth Center President-elect, to catalyze and harness these changes, as he eloquently described to the Danforth Center faculty this week. In that meeting, Giles also stressed what should not change - our mission, vision and values – and that should give everyone optimism about the Center’s future.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Much of the lunchtime discussion and hallway chatter these days concerns the disruptive actions that put federal research funding at risk, or that affect how we support a diverse, inclusive community at the Danforth Center. I’m a part of many such conversations. In this week’s message, however, I want to highlight several positive developments or achievements at the Center.
Three new NSF grant awards. After short delays, we were thrilled to receive award notices this week for three new grants from the National Science Foundation. Despite considerable uncertainty across the funding agencies, NSF has restarted issuing awards for competitive projects. Let’s hope this foreshadows a restart of granting activity at other agencies that are considering our proposals. Congratulations to the Eveland, Miller and Burch-Smith/Czymmek teams for their grant-getting success!
Affirmation of commitment to a diverse community. At the Faculty Meeting last Monday, we had a good discussion about how we intend to support our diverse Center community in light of recent federal restrictions. The good news is that the Danforth Center remains fully committed to being an inclusive and welcoming place, recognizing strength through our differences and embracing contributions from all who join our community. We may not receive financial support for “broader impacts” that have traditionally been supported (and often required!) by various funding agencies, but our values remain intact and unwavering.
Progress with field site construction. Are you following progress with facilities development at the Danforth Center Field Site? Terry Beeler is regularly posting photos of progress on construction of the field site Pavilion and Building E on Slack (# DDPSCfieldresearchsite). The Pavilion will meet human needs for those working at or visiting the site, while Building E will serve as a plant processing and working facility for those running experiments. Thank you, Terry, for enabling us to see the field site develop!
Nice work from Mohammad Fazle Azim. A wonderful example of the good work happening at the Danforth Center every day was given by Mohammad at this week’s mentoring seminar. Mohammad, originally from Bangladesh, is a nearly finished Ph. D. student in Tessa Burch-Smith’s team, and his thesis research focuses on how information is transmitted between plant cells. His careful work has relevance to how plants control their growth and how viruses move from one cell to another. Well done, Mohammad!
These highlights do not erase the challenges we face, but they’re a good reminder to remain focused, control what we can control, and stay committed to the work we’re here to do.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
“Fear defeats more people than any other thing in the world.”
– Ralph Waldo Emerson
To follow on from my message last week, many Danforth Center community members have been stressed by recent executive orders from the White House. The language in one order suggested that federal grant funding for domestic research, fellowships, student aid and other programs would be paused or suspended, which could have a significant impact on the Center. Some degree of relief was found when this order was rescinded shortly after it was issued, due at least in part to strong objections and challenges from individuals and grant-receiving organizations across the country. On Monday, the Center’s views about the importance of federal grant funding to the Center and the state were communicated to our elected representatives at both the national and state levels.
To date, we have received official notice from only one agency about suspension of activity on a funded project. This suspension was not related to the order that was subsequently rescinded, but rather to a separate executive order about reevaluating and realigning foreign aid. The suspension is manageable in the near term, but how long this will last is not clear. Whether or not new executive orders will be issued to slow down, pause or threaten grant funding from other federal agencies is not known. My sense is that it will be extremely difficult to reduce or eliminate funding that Congress has appropriated for scientific research to domestic organizations like the Danforth Center, but I would also expect delays in proposal processing and award notices due to disorder that has been injected into the federal grant-making systems.
I ask that members of the Danforth Center community be patient during this time, despite the external events and agitation. I also ask that we continue to focus on our work and those things we can control, including submission of new grant proposals that teams have worked so hard to prepare. We are an organization with the will and capacity to deal with some turbulence, if it occurs.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Changes of administrations by newly elected U.S. Presidents always trigger concerns about federal government priorities and continuity of funding for agencies and programs that we care about. It’s fair to say that those concerns might be elevated during this cycle. As a research organization with significant funding from several federal agencies, the Danforth Center has a particularly keen interest in continuity of funding that supports competitive scientific research.
In 2024 the Danforth Cetner was awarded $20.7M in overall research grants and contracts, of which nearly 58% was from federal agencies. With 31% of that funding, the National Science Foundation (NSF) was the frontrunner among federal sources, followed by the Department of Energy (9%) and Department of Agriculture (7%). The Bill & Melinda Gates Foundation (12%) was the only non-federal organization within the top four funders of grants to the Danforth Center. When crafting the 2025 budget, we anticipated similar trends with research grants and contracts accounting for roughly half of the overall budget. Continuity of funding from federal granting agencies is really important to us!
Historically, support for research granting agencies by Congress has been bipartisan, with notable leaders from Missouri. When he was chair of the Senate subcommittee with responsibility for Health and Human Services, former Senator Roy Blunt led the increase in appropriations for the National Institutes of Health budget by roughly 50% between 2015 and 2022. In 1998, former Senator Kit Bond of Missouri was the champion for creation and funding of the NSF Plant Genome Research Program, which has funded numerous past and current Danforth Center projects.
These and other agencies that award grants to the Danforth Center have a big impact on St. Louis, the state and the nation. Federal funding has enabled the Center to grow as a hub for scientific talent, exceptional facilities, and collaborative research with both public and private partners. Counting the Danforth Center, BRDG Park on our campus, and the Helix incubator next door, we have an estimated annual economic impact of $480M on the St. Louis region alone.
During this transitionary period, we are building awareness with elected officials about the critical role of federal grant funding for the Center, the growth of St. Louis, and our ability to achieve impact. The exceptional work done by Danforth Center community members means that’s a compelling case to make!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
In an open letter signed by over 150 Nobel Prize and World Food Prize winners, the critical need to address the “tragic mismatch between global food supply and demand” was published recently. Organized by Cary Fowler, a 2024 World Food Prize Laureate, the letter makes an urgent plea for both financial and political support for environmentally responsible and climate-compatible“moonshot” technologies to “transform our food systems to meet the nutritional needs of everyone sustainably” over the next 25 years. The letter emphasizes that 700,000,000 people worldwide are currently food insecure, with the most vulnerable populations in regions that are most affected by climate change, soil erosion and other environmental challenges that threaten food production.
As Allison Miller pointed out yesterday, the last paragraph of the letter lists examples of research investments in a food security “moonshot” that are needed. Noting unparalleled recent advances from genome editing to AI, the authors highlight the following areas in which to concentrate:
“…enhancement of photosynthesis in crops such as wheat and rice; biological nitrogen fixation of major cereals; transformation of annual to perennial crops; development of new and overlooked crops; innovations in diverse cropping systems; enhancement of fruits and vegetables to improve storage and shelf life and to increase food safety; and the creation of nutrient-rich food from microorganisms and fungi. Also critical will be the study and development of strategies to make certain that the fruits of these scientific research initiatives reach and benefit those most in need.”
Those who have been paying attention to Danforth Center priorities, discoveries, and investments will recognize most the areas highlighted. In fact, the letter could not be more aligned with the mission and vision of the Danforth Center, and with where we see the need for current and long-term investments in research and development. Providing accessible, affordable and nutritious food in nature-compatible ways around the world is a massively complex problem involving social, political, and economic factors. But at its core, this is a technological challenge that needs expanded and continued scientific effort and support, and different ways of thinking. The Danforth Center will continue to grow in relevance. And I look forward to seeing the Center’s impact grow as well in the years ahead.
P.S. Cary Fowler is scheduled to be the Center’s Seeds of Change speaker on Wednesday, August 27, 2025.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Danforth Center scientists publish over 100 research articles describing discoveries and ideas each year. These are each important team and research program milestones. The problem is, most articles are published in highly technical scientific journals, meaning they are largely beyond the grasp of scientists outside of the authors’ fields and the public. My periodic synopses of selected, recent articles are intended for everyone to better understand and appreciate these important contributions by Center scientists and their collaborators. Let’s have a look at three more today.
Bertolini E, Manjunath M, Ge W, Murphy MD, Inaoka M, Fliege C, Eveland AL, and Lipka AE (2024). Genomic prediction of cereal crop architectural traits using models informed by gene regulatory circuitries from maize. Genetics, doi.org/10.1093/genetics/iyae162.
The Eveland team has been analyzing the networks of genes that control plant architectural traits, like leaf angle and branching patterns. These are critical traits because they determine how dense a crop can be planted; densely plant corn, for example, enables higher productivity per acre. Here, they used predictive modeling with extensive genomic and phenotypic data to reveal similarities or differences between the networks controlling architecture in maize and other plants, including sorghum (similar) or rice (different).
Nboyine JA, Umar ML, Adazebra GA, Utono IM, Agrengsore P, Awuku FJ, Ishiyaku MF, Barrero JM, Higgins TJV, and MacKenzie DJ (2024). Assessment of field performance and bruchid resistance during seed storage of genetically modified cowpea expressing the alpha-amylase inhibitor 1 protein from common bean. Frontiers in Plant Science, doi.org/10.3389/fpls.2024.1478700.
Don MacKenzie and collaborators are seeking to develop new varieties of cowpea with resistance to a destructive post-harvest pest, the cowpea weevil, which can cause 100% loss of grain in storage. The team introduced the alpha-amylase 1 protein from common bean into cowpea, and in field and post-harvest trials showed that the resulting plants were remarkably resistant to weevil infestation for at least four months in storage. Combined with resistance to cowpea pod borer, which the team brought to market in Nigeria and Ghana, this advance paves the way to addressing two of the most significant cowpea pests in West Africa.
Godwin J, Djami-Tchatchou AT, Velivelli SLS, Tetorya M, Kalunke R, Pokhrel A, Zhou M, Buchko GW, Czymmek KJ, and Shah DM (2024). Chickpea NCR13 disulfide cross-linking variants exhibit profound differences in antifungal activity and modes of action. PLoS Pathogens, doi:10.1371/journal.ppat.1012745.
The Shah and Czymmek teams continue their productive collaboration on small anti-microbial peptides from plants to understand factors that contribute to potency and mode of action against fungal pathogens. A chickpea peptide, NCR13, was found to adopt two different structural conformations that differed greatly in antifungal activities, leading to key insights that inform how to design and produce more effective and sustainable alternatives to chemical fungicides.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I have never made a New Year’s resolution. The logic of waiting until January to either start or stop doing something to improve oneself never made sense to me. On the other hand, I’m a strong believer in defining and setting goals for the new year. During all year-end professional development conversations in which I participate at the Danforth Center, more time is spent on goal-setting than on any other topic. Here are three thoughts that I’ve found to be particularly helpful for setting and delivering on goals through the years.
Goals should focus on achievement. While that might seem rather obvious, there is some nuance to consider. The process of setting goals should start with clearly defining what you want or need to achieve. Anchoring goals to desired achievements or outcomes serves to instill purpose, direction, clarity and focus. Goals that are untethered to desired achievements risk being focused on less important activities. Which reminds us of an insightful quote from the late, legendary coach, John Wooden: “Never confuse activity for achievement.”
Goals should reflect priorities. Not only should goals be focused on desired achievements, but they should reflect the priorities of the team. Priorities, or the most important functions and desired outcomes, should dictate where team members’ effort and time, funding, and other resources are invested. In my case, annual goals are typically linked to the six prioritized outcomes as articulated in the Center’s 2021-2025 Strategic Plan.
Goals will help you say “No.” Did I just hear someone gasp? “No” can be a harsh, alienating, and improvisation-killing word. Yes, and, saying “no” is essential for getting things done, keeping the team focused, and staying on track. In a creative environment like that Danforth Center, there is frequent pressure for teams to start under-resourced side projects, to accept unfunded invitations to collaborate, or to do more than what capacity allows. For leaders, having well-defined, priorities-centered goals to which resources are already committed will help when the time comes to respond with an appropriate form of “no.” In fact, I keep a printed copy of annual goals on my office desk at all times, in part as a reminder for this purpose.
Many thanks to all who have set meaningful, thoughtful goals for 2025, and to everyone else, there is still time. I wish everyone a productive and rewarding 2025!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
With the holiday rapidly approaching, I want to give everyone the gift of brevity with this final message of 2024. Looking back on the year, I am proud of all who contributed and achieved to move the Danforth Center forward and deliver on our mission. Some of you were doing so with high visibility, while so many others were vitally important behind the scenes. It all matters greatly! Looking forward to 2025, we have some transitions to anticipate, but they are all happening from a solid, stable base that we’ve worked hard to build together as a Center community. I’m excited to see everything that is just now on the horizon come into view in the new year.
Thank you for all you’ve done this year, and I wish you the very best of Holiday Seasons.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
As we do each year, we recently celebrated milestone anniversaries for Danforth Center community members who have been with us for 5, 10, 15 and 20 years. Talented individuals have many opportunities, so the fact that so many chose to stay with us for long times during their careers is meaningful. At several events this week, I asked 10 individuals who have been at the Danforth Center for at least five years, “Why have you chosen to stay as long as you have?” Here are the most common responses I received.
- Strong connection with the mission. This was the first answer that nearly everyone gave! Identification with the Center’s higher purpose comes through in every interview and survey we do. Several individuals this week offered striking comparisons to prior jobs at organizations that lacked a compelling mission, stating that this was the first feature that attracted their interest in the Danforth Center.
- The people. “I love the people I work with” was the second most common response. Several individuals with administrative roles mentioned they how much they like working with the scientists. One made special reference to the young scientists and the energy and excitement they bring to the Center.
- Alignment with professional aspirations and needs. Many of the scientists I spoke with mentioned this, and for good reason. If you have sights on achieving great things in plant science, the Danforth Center is built for you! But several non-scientists also mentioned alignment with career goals and the interesting nature of their work. One person mentioned the importance of offering flexible or hybrid work arrangements in their department.
- Ability to grow and make a difference. Hal Davies (VP for Finance and COO), who will retire soon after more than 23 years at the Center, was eloquent in expressing reasons for his long tenure. It wasn’t just because he could ascend to new roles and exercise his business and financial skills. He gained the most satisfaction by helping the Danforth Center and its people deliver on the mission, while also stewarding the financial resources with which we’ve been entrusted.
In study after study, these four themes emerge as important, though not the only, reasons for long-term job satisfaction in a wide range of fields and industries. Many thanks to all who were recognized with milestone anniversaries this week, and to the thoughtful, long-standing community members who participated in my informal survey.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
In remarks during her acceptance of the Danforth Award for Plant Science yesterday, Toby Kellogg spoke eloquently about the value of basic scientific research, which can be defined as research aimed towards better understanding of the natural world. Traditionally, basic research is contrasted with applied, or problem-solving, research that focuses on specific, practical solutions. Seeking to understand separated oscillating fields would be considered basic research in physics, while research for the purpose of improving satellite communications would be considered applied. But the problem with this basic vs. applied distinction is, it’s an illusion! Without research on separated oscillatory fields, for which Norman Ramsey and colleagues won the Nobel Prize in 1989, development of practical satellite communications would not be possible. The basic and applied distinction does not work well for me.
In Toby’s case, her work to understand natural diversity and evolution of the grasses was considered basic research, that is, until a few decades ago when plant genome sequencing teams realized they needed Toby to understand the new data. Toby and her lab team of evolutionary biologists provided key insights to interpret the complexities of plant genomes, which contributed vital information for breeders in crop improvement programs. When I ponder Toby’s contributions, I think about how she’s helped us understand the natural world of plants, and how she’s enabled countless crop breeders to work more productively and quickly.
I see the active integration of basic and applied research every day at the Danforth Center. In a Gates Foundation-funded project, for example, we’re seeking to develop and improve cassava traits for breeders and smallholder farmers through an entirely new approach: targeted epigenetics (if you trust me here, I’ll spare you the details of epigenetics!). Collaborating teams at the Danforth Center, UCLA and the University of Hawaii are discovering new epigenetic mechanisms in plants and applying them to make disease-resistant cassava. It’s a collaboration that does not distinguish between basic and applied contributions.
Basic research provides the foundation on which application-directed research is done. But like a house, why consider the foundation as something separate from everything else? Setting a distinction between basic and applied research, or valuing one more than the other, are pointless exercises. That basic idea certainly applies here at the Danforth Center.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
For the past few years, I’ve started preparing a Weekly Message with a Thanksgiving theme to send on the Wednesday before the holiday. And each year, I’m interrupted by Anna Dibble (VP, People and Culture), who sends me a message like: “Hey Jim, I am planning on sending out a Thanksgiving message to the community today…forgot to chat about that with you yesterday.” Then I read what she’s planning to send, and it’s always incredible! It’s heartfelt, warm, and sincerely appreciative towards the Danforth Center community, in the finest tradition of Thanksgiving messages. I then get back in touch with Anna with a message to the effect of, “This is so good, how do you expect me to follow that?” Upstaged…again!
I cannot match Anna’s message, and that’s part of what I’m thankful for this year. I’m thankful that we have so many talented people, like Anna, who are investing so much of themselves to contribute to the Danforth Center community. I’m thankful to work with so many who identify with our mission and work together to make the Danforth Center entirely unique and impactful. And I’m thankful for the determination and passion you show to get through the difficult moments, or to rise above the skepticism from others who think you can’t do it.
I hope you read and think about Anna’s message from this morning, and then pass along your appreciation and gratitude to others who make a difference for you at the Danforth Center. Happy Thanksgiving!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Across the Danforth Center and most other organizations, there is constant tension between innovating and getting things done. It’s highly likely that there are better ways to operate in each of our research groups, teams and departments to increase the quality, efficiency or relevancy of our work. But developing, testing, learning and implementing different, innovative ways of operating is difficult when resources and effort are already maxed out to deliver on what is expected or needed. How do we innovate to get better, while also minimizing the impact on getting things done in the meantime?
Consider the idea of a “space to innovate” to test and refine new ideas and technologies without interfering or competing with ongoing work in the near term. This means much more than simply setting aside a lab or distinct physical workspace. It also means creating financial space in the budget to cover the costs, space in the strategic planning processes, time space, and space to fail. The space to fail is critical, as learning from failure is an essential part of the innovation process.
We’re serious about this space to innovate idea, which is why we created the Plant Biotechnology Innovation (PBI) Hub earlier this year to design, develop and test better systems for plant transformation and genome editing. Most of our research teams have critical needs for better, publicly accessible methods to edit and regenerate plants, for both fundamental research and crop improvement purposes, but they lack resources to develop those improvements within their labs. The PBI Hub team of Ashley Snouffer and Marissa North has already worked productively with nine research teams to develop improved methods and tools for nine plant species, including cassava, tef and tomato. Innovations developed in the PBI Hub can be brought back to individual labs, or transferred to the Plant Transformation Core Facility for scale-up and production. Many thanks to Ashley and Marissa, and all of the collaborating teams, for creating and developing the PBI Hub!
There are other good examples of research teams and core facilities creating space to innovate. And with careful planning, teamwork, and communication, I see possibilities to create space to innovate within administrative teams as well. As a research institution with Innovation as a core value, creating such spaces across the Center makes a lot of sense to me.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
With the recent U.S. elections on many minds, I want to underscore a vital strength of the Danforth Center: We are a non-political organization working in a non-partisan manner to benefit people regardless of their beliefs. If a community member, collaborator or supporter is willing to help deliver on our mission and realize our vision, and do so in a way that aligns with our values, we welcome participation by those who hold views from across the political spectrum. The “science” part of our mission – to improve the human condition through plant science – requires us to remain objective, data-based, and apolitical in our work. To do otherwise would degrade our credibility and impact, and fracture support or perceived relevance along ideological lines. Despite forces that may seek to politicize some issues that we’re addressing, I have few worries that we’ll stray from what we should be.
But I worry about how these challenging and polarizing times, and the recent campaigns and elections, are affecting community members at the Danforth Center. Research by the American Psychological Association (APA) shows that the vast majority of adults are negatively affected by the polarization, with 77% feeling significant stress about the future of the nation. The specifics underlying why people are stressed may vary depending on their strongly held beliefs, but the resulting stress cuts across political boundaries. The Danforth Center community is not immune, as most of us have experienced recently. “Tired,” “despondent,” “disappointed,” and “exhausted” are one-word descriptions to summarize how four of our colleagues feel today. At the same time, I’m encouraged by how these colleagues and others are helping stressed team members cope, like through regular check-ins and one-on-one meetings to hear concerns.
While I’m concerned about the well-being of community members, I’ve seen little evidence that the Danforth Center has become a more polarized place. Our community members hold differing political affiliations and beliefs, yet we see relatively few problems because to those differences. I believe this is due, at least in part, to the fact that we have strong identify with a unifying, non-partisan mission, and to the fact that we value getting to know one another. The APA notes that interacting regularly with those who hold different beliefs from your own has the great benefit of revealing shared experiences, values and aspirations, and that builds better understanding and appreciation of one another.
To everyone who seeks to understand and help your Danforth Center colleagues, I thank you. We are stronger as a result. And for Danforth Center community members and dependents who may need help from professional counselors, our Employee Assistance Program is available to you at any time (information on Workvivo in People and Culture space).
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
At the Danforth Center’s Seeds of Change event this week, Penny Pennington presented us with an idea: Innovation and positive change starts with falling in love with a problem. Along with a terrific panel of Allison Miller (Danforth Center), Meredith Malone (Curator, Mildred Lane Kemper Art Museum), and Christine Corday (multidisciplinary artist), Penny explained how passion for a problem leads to better teams, more breakthroughs, and more impact. Like many others who attended, I’ve continued thinking about this idea since the event.
First of all, I very much agree with Penny’s thesis. Can anyone think of an individual who made a groundbreaking discovery or developed a great product who did not love the problem they were seeking to solve? I cannot. The late Steve Jobs built one innovative, transformative product after another when he led Apple, and he described on many occasions how this was only possible because he and his team truly loved the problems they were solving and the vision of what they could achieve. In one interview, Jobs described how difficult it is to build something new, and that success involves perseverance, setbacks and worrying over long periods of time. He explained that if you don’t love what you’re doing, you’ll probably give up and go do something else.
We’ve all heard the overused clichés, often from commencement speakers: “Do what you love,” or “Find your passion and go for it!” I think the sentiment often being conveyed by these statements is, do what you love because that will make you happy. I’m not convinced that’s the true value of loving the problem you’ve taken on. Your passion is what gets you through the dead-ends, the misery, the failures and the criticism that will undoubtedly come your way. Love for the problem will keep you in the game, despite the intensity of the challenges. Rather than happiness, I suspect that passion for the problem ultimately results in more fulfillment of purpose and satisfaction that you’ve made a positive impact.
Scientific research and all the work to make the Danforth Center run well are difficult endeavors. But every day, I see so many purpose-driven Danforth Center community members who love the hard problems they’re addressing and the prospect of positive impact when they achieve success.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
The most fun I had this week was participating as a judge in an Elevator Pitch competition at the BioBash event organized by CSTM. With fellow judges Becky Bart and Bethany Zolman (Professor of Biology, UMSL), and a well-attended BioBash audience, we heard one-minute presentations from 10 early career scientists, ranging from two undergraduate students at Missouri S&T University to postdoctoral scientists at the Danforth Center. The speakers’ objectives were to introduce themselves, describe what they do and why it’s important, and communicate what makes them unique. In other words, they needed to give the elevator pitch about themselves and their purpose.
A great elevator pitch needs a great opening – a hook – to immediately capture the interest, imagination and curiosity of the listener. And we heard some great starts! Before telling us why microscopic imaging is so important, Samantha Nuzzi started with, “I’ve seen things you’ve never seen!” Marisa North explained how she embraces adapting herself to unique professional situations, but she first said, “I’m a Transformer” (think Optimus Prime). Neeta Lohani began with, “I’m fascinated by the soybean family tree,” to communicate about her goal to help develop more resilient crops and sustainable agriculture.
A great elevator pitch needs to engage the listener on their terms, and in their language. I was impressed with how the speakers erased coded language unique to their specialties, or used creative devises to bring their point of view to life. Using a wonderfully simple graphic to illustrate that science solved the ozone hole problem, Stewart Morley effectively stated, “Science can solve the climate change crisis, too!” Balaji Balamurugan illustrated nicely how his experience, skills, strengths and purpose define his career aspirations.
A great elevator pitch also needs to be concise, with the speaker attentive to the reality that elevator rides are really short! Brief presentations are the most difficult and require the most preparation, and I admire the work each presenter put into their pitch. I also admire the courage of each speaker in presenting their personal stories.
Congratulations to the judge-selected winners, Samantha Nuzzi and Neeta Lohani (each from Danforth Center), to the audience-selected winner, Jeremy Howard (UMSL), and to all of the presenters who each took us on a wonderful ride!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Believe it or not, the 2025 budget process ongoing now at the Danforth Center is an interesting process! It starts with each department leader preparing a proposed budget with all projected expenses and revenue for the coming year, along with brief summaries of their team, recent achievements, goals, opportunities and challenges. Each department leader meets with the team of Djuan Coleman (Director of Financial Planning), Hal Davies (VP for Finance), Denise Coffer (Manager of Financial Planning) and me, during which we ask lots of question and provide guidance for budget revisions. After all department budgets are presented, the finance team pulls together a Center draft budget and we ask the following questions: Does this budget support Center priorities, needs and goals, and will we have the money to pay for everything? If one or both answers is “No,” then we have more work to do before seeking approval of the budget from the Board of Directors in November.
The budget process focuses my attention on three critical areas: 1) Our mission and strategic priorities; 2) our capabilities and capacity; and 3) our financial sustainability. The work we do at the Danforth Center should fall at the intersection of all three areas, outside of which is problematic. For example, a proposed project that is mission-aligned and within our capabilities, but lacking a source of funding, can’t get very far. A program that is mission-aligned and somehow funded, but outside of our capabilities, is a bit delusional. This simple guide helps us make decisions about resource allocation during the budget process.
This year, I’ve taken note of how many core facilities are delivering on needs, growing efficiencies, and bolstering their financial positions. The Data Science Core has grown data analysis services that benefit more research teams, but with lower costs due to partnerships with other cores and labs, migration to cloud computing/data storage, and better alignment of team members’ capabilities with the actual needs. The Bioanalytical Chemistry Facility (formerly known as Proteomics and Mass Spectrometry) has expanded services, increased numbers of users, and generated more revenue by consolidation with the Ionomics team and cross-training of team members. The Plant Transformation Facility has been able grow revenue to offset costs, but without increasing the size of the team. These kinds of developments help everyone because they relieve pressure on the overall budget.
Thank you to Djuan, Denise, Hal and all of the department leaders who are working hard to deliver a manageable, purposeful budget for the Danforth Center.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Yesterday morning, Stephanie Regagnon let me know that she will be leaving her position as Executive Director of Innovation Partnerships at the Danforth Center in December, 2024. Stephanie has been an important part of the Center for over four years, joining us at a difficult time early in the COVID-19 pandemic. Today, I want to recognize Stephanie’s achievements and the mark she’s left on the Danforth Center.
Before taking her position, Stephanie was already heavily invested in the Danforth Center as a volunteer on the Friends Committee. She was particularly excited about our vision for growing and strengthening the St. Louis region through plant science. Her long experience in the agricultural industry made her an excellent choice for the newly created position to lead the Innovation Team, with responsibility for intellectual property management and innovation partnerships. She contributed early to the 2021-2025 Strategic Plan, which articulated goals for a Start-Up Initiative and laid the conceptual foundation for creation of Danforth Technology Company to spin out new companies from the Center.
Stephanie solidified existing partnerships, like with the St. Louis Community College, and developed new partnerships to advance BRDG Park and the broader innovation district, 39North. Among other achievements, she helped recruit or retain several companies in 39North. Until recently, the Danforth Center was the primary organization providing support for the district. Stephanie helped change that when she worked closely with regional partners to define the need for an independent organization to run and elevate 39North, which came to life in 2023. Stephanie serves as a board director for 39North.
Stephanie and team have also worked passionately to grow the geospatial industry here, build up a more equitable and well-trained workforce in the region, bring people together, and create international connections between AgTech organizations in St. Louis and Latin America. She is a very familiar presence around St. Louis, and has represented the Danforth Center enthusiastically on countless occasions.
The Danforth Center has changed in many ways over the past four years, and Stephanie Regagnon has had a productive hand in that happening. I sincerely thank Stephanie for all of the work she’s contributed toward our success!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Most weeks at the Danforth Center, there’s at least one visiting speaker who presents a formal presentation of their research. This week, we invited Steve Fiore (University of Central Florida), a social scientist who studies how trans-disciplinary teams work together. Understanding what distinguishes successful teams from others is a big deal, especially for the Danforth Center, as nearly all of our work is done in collaborative teams with members coming from diverse disciplines. Here are a few insights from Steve’s fascinating work and presentation.
Successful teams remember their ABCs – They pay close attention to Affective (or attitudinal), Behavioral (or skills), and Cognitive (or knowledge) components of their team member interactions. That is, successful teams have members who are attentive to one another’s well-being and needs, go out of their way to understand their colleagues unique perspectives, and productively use team member’s diverse knowledge and experience. Successful teams are not afraid to acknowledge members’ strengths and weaknesses, and to allocate responsibilities that align with individual talents. Successful teams take the time to craft a mission or purpose, around which they align with common goals.
Successful teams tolerate more, and learn from, mistakes – This results in part from the fact that successful teams encourage riskier ideas, and those may lead to more errors. But when mistakes are made, successful teams are better at learning from the experience and adjusting. The key is not to avoid mistakes, but rather to treat mistakes as normal occurrences and use them productively. As one with vast experience in making mistakes, this point really resonated with me. The company Google has taken this further and institutionalized a formal process to detect, diagnose and learn from mistakes without assigning blame or penalty.
Successful teams promote psychological safety – Members of successful teams promote everyone’s input and insights, regardless of hierarchy or status. In other words, successful teams provide a safe place for everyone to offer critiques, point out mistakes, and challenge ideas without fear of repercussions. This promotes more contributions from early-career team members, who often possess more knowledge about, say, modern techniques than the seasoned team members. Promoting psychological safety, Steve pointed out, involves behaviors like active listening and demonstrating empathy towards those team members who may feel vulnerable.
This presentation was not our typical “plant science seminar,” but I cannot recall another from which we can all learn more.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
As a not-for-profit organization, part of our operating budget is funded by donations from individuals and families, foundations, and companies that generously support our work. Some donors may give for specific, designated purposes, like to support a graduate student fellowship or the Proof-of-Concept program. Others may not want their gifts designated for a singular purpose, but instead, directed to support work within a range of priority areas depending on where we have pressing needs. That’s why you should know about the Danforth Center Impact Fund.
The Impact Fund pools gifts from many donors, including those who give annually, to support Danforth Center priorities in four areas (allocation percentage indicated):
- Early-Stage Projects and Research (55%) – This helps get new Danforth Center labs up and running when we recruit a new faculty member, and it provides seed funding to test interesting ideas. This enables preliminary work to be done so that subsequent large project proposals to funding agencies and foundations are competitive.
- Training and Internships (22%) – The Impact Fund enables us to fund more undergraduate Summer interns, and to support the annual CSTM budget. Assuring funding for graduate students from university partners has enabled growth to nearly 40 students over the past year.
- Education and Outreach (15%) – This enables a base of support for education and outreach to maintain continuity of K-12 student programs and teacher training. Partial funding for the core team in Education Research and Outreach Laboratory (EROL) enables collaboration with other Danforth Center lab teams for broader impacts.
- Equipment, Technology and Facility Improvement (12%) – The Impact Fund supports acquisition of shared laboratory equipment and investments in Core Facilities. It enables adaptation of facilities for new purposes when needs change, and flexibility that is needed as technology evolves.
Most of the funding needs in these areas cannot be met by traditional grants or other mechanisms, meaning that the Impact Fund serves a unique role in supporting priorities at the Danforth Center.
Over the past year, approximately 1,100 donors, including many of you, supported our work with gifts via the Impact Fund. And tonight, proceeds from Party With the Plants will benefit the Center through the Impact Fund. I give thanks to the Development team that works so productively in stewarding our supporters, and to everyone who chooses to give generously to the Danforth Center!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I recently saw a group of attendees at a scientific conference we hosted at the Danforth Center looking at the mounted plaques for past recipients of the Danforth Plant Science Award. Visiting scientists frequently stop at the wall commemorating prior award winners and find recognizable names of diverse individuals who have made a big difference in plant science. The criteria used to select Danforth Plant Science Award recipients are fairly simple – exceptional scientific achievements through their research, or exceptional impact in agriculture, food and nutrition, or human health. Some past awardees, like Norman Borlaug or Joanne Chory, are the equivalent of “household names” in plant science. Some others, like Ethiopian scientist Segenet Kelemu, have had impact that vastly exceeds their name recognition.
Like so many past recipients, the next Danforth Plant Science Awardee is someone who has made an indelible mark in science and our understanding of the natural world. She built a career of scientific achievement by asking the simplest of questions like: How and why are these two plants different? Those fundamental questions are relevant to understanding how crop plants evolved from wild relatives. Or to understanding why mature seeds of some plants drop to the ground and others do not. Or to acceleration of breeding efforts for short-stature tef that withstands heavy rain and wind. This year’s recipient is a deeply insightful scientific leader who has made fundamental discoveries about how plant species change over evolutionary time as their environments change, and how they changed for human benefit over the course of crop domestication. And to top it off, she’s one of the Danforth Center’s more admired scientists!
This year’s Danforth Plant Science Award recipient is the one and only Elizabeth (Toby) A. Kellogg. Toby is the Robert E. King Distinguished Investigator and Member, Danforth Plant Science Center, since 2013. She’s been a guiding light within the plant science community for decades, and an entirely influential colleague for all who work with or learn from her. This award both recognizes Toby’s stellar achievements in science, and thanks her for the countless contributions she’s made to the Danforth Center.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
When looking around, I see countless examples of contributions to the Danforth Center community. “Contributions to community” is a term used to describe work we do that is beyond our specific jobs and roles, but that benefits the broader Center community. None of the CSTM co-chairs have to be CSTM leaders, but they contribute to community by serving their colleagues. None of the teams that build displays and interact with visitors at PlantTech Jam or Party with the Plants has to spend time and effort in this way, but they contribute to community through communication about our work and making the Center shine. I appreciate how contributions to community, like these examples, make the Danforth Center a better.
But contributions to the Danforth Center community are not limited to those of us with jobs here. Contributions of volunteer time and effort from across the region and beyond are numerous, and vital to our success. Service on our Board of Directors or other volunteer groups, like the Friends Committee or tour docents, provides governance functions, connectivity with prospective donors, and representation of the Center to the public. The late Jim Knight was one of the earliest such volunteer contributors; we were so appreciative that we created the Newell S. “Jim” Knight, Jr. Volunteer Award to recognize exceptional service. Besides Jim as the inaugural awardee, we have recognized George Fonyo, Jim Johnson, and Molly Cline with this award.
And just yesterday, we were thrilled to honor Ruth Kim as the fifth awardee. Ruth has served and represented the Danforth Center in so many ways, including as a current, highly active Director on our Board and a past member of the Friends Committee. She gives her time, effort and resources generously, but that’s only part of her story. She has introduced hundreds of her friends and associates to the Danforth Center through invitations to lunches, receptions and events like those at which our community member contribute. Ruth’s enthusiastic efforts have grown our base of donors and elevated awareness of the Danforth Center. Along with Jim Knight and past award recipients, Ruth is passionate about the Center’s mission and has achieved superhero status within our remarkable volunteer community!
Thank you to all who contribute to the Danforth Center community in ways that make a difference. And congratulations to Ruth Kim for her extraordinary achievements in elevating the Danforth Center.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Diving deep into a scientific problem always results in surprising discoveries, detailed mechanistic understanding, new and important research directions, or unexpected applications that address real needs. Let’s highlight and briefly explain three recent publications, and recognize three Danforth Center teams that have persisted in understanding three different areas of plant science.
Transposase-assisted target-site integration for efficient plant genome engineering.
Liu P, Panda K, Edwards SA, Swanson R, Yi H, Pandesha P, Hung YH, Klaas G, Ye X, Collins MV, Renken KN, Gilbertson LA, Veena V, Hancock CN, Slotkin RK. Nature. (2024) doi: 10.1038/s41586-024-07613-8. The Slotkin team has made numerous, insightful discoveries about how plant transposons, which can jump from place to place in a genome, are either locked in place or enabled to jump. From that deep understanding, the team developed ways to harness the unique attributes of certain transposons to insert new DNA into specific, desired sites in the genome. This opens up vast possibilities to accelerate crop improvement in ways that increase precision and lower costs. You have not heard the last about this new technology!
SymRK regulates G-protein signaling during nodulation in soybean (Glycine max) by modifying RGS phosphorylation and activity.
Choudhury SR and Pandey S. Molec. Plant Microbe Interact. (2024) doi: 10.1094/MPMI-04-24-0036-R. The Pandey lab has focused intensely on how plants respond to changes in their environment, like when certain symbiotic bacteria interact with roots. Legumes like soybean interact with specific beneficial bacteria (e.g. rhizobia), resulting in nodules that churn out a natural form of fertilizer for the plant. The team here revealed several new, critical molecular interactions involving G-proteins and receptors that enable plants to engage with the bacteria.
pyMS-Vis, an open-source Python application for visualizing and investigating deconvoluted to-down mass spectrometric experiments: A histone proteoform case study.
Pesavento JJ, Bindra MS, Das U, Rommelfanger SR, Zhou M, Paša-Tolić L, Umen JG. Analytical Chem. (2024) doi: 10.1021/acs.analchem.4c02650. The Umen team is well known for longstanding, elegant research to understand how algal cells multiply and differentiate into sexually distinct types. This paper revealed not only new insights into how algal genomes are controlled during the cell cycle, but also an entirely new method that any other team can use to measure and compare different forms of proteins and protein complexes.
I admire the persistence and commitment needed to solve hard scientific problems. And I sincerely appreciate when that effort yields important insight and application beyond what was initially envisioned!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Around 12 years ago, I was sitting next to the late Jack Taylor at a dinner. In 1957, Jack founded and built the company that was later renamed Enterprise Rent-a-Car, which ultimately grew into the seventh largest private company in the U.S., Enterprise Mobility. I knew he had cultivated a great company culture when he was CEO, so I asked Jack, “How did you did you do that?” His initial reply was unsatisfying. “I don’t know, I never understood all of that business culture stuff,” he said. I pressed him some more with, “Seriously, you must have been doing something right.” Then he laughed and said, “I don’t know what I did. I just came in every morning and asked people, ‘Are you having fun?’”
The data are in on the topic of fun in the workplace. In their book, “Work Made Fun Gets Done,” Bob Nelson and Mario Tamayo studied the differences between organizations that do or do not rank among the 100 Best Places to Work (based on employee ratings) as published in Fortune. Eighty-one percent of employees at organizations that made the list described their workplace as fun; at organizations that did not make the list, that number was 31% lower. Research indicates that fun at work promotes engagement and creativity, increases productivity and resilience, and lowers employee turnover.
Encouraging workplace fun doesn’t mean we have a license for disruptive pranks or hazardous horseplay. But it does mean different things to different people. Nelson and Tamayo point out that fun can be gained by turning routine tasks into games or contests (e.g. first to finish). I like little competitions among coworkers, like guessing how many people will show up in person for a seminar presentation. Some find music in the workplace, like in the Café or at Wednesday morning tea times, adds a fun element. Granting a little time for fun, like 40 minutes to compete in Lab Olympics or laughing with colleagues during a work-free lunch, can make a difference. Even our public events, like the upcoming Party With the Plants, have much room for fun engagement with one another.
Jack Taylor found a way to see humor and have fun in most situations, at least those in which I interacted with him. I suspect his sense of fun was infectious, and served as a major ingredient in his company’s success. I think that’s worth emulating.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
At a large gathering of Danforth Center supporters last evening, I shared one of the best things about being a scientist who makes a discovery. For some amount of time – a few minutes, a few days – you are the only person in the universe who knows that particular bit of knowledge. If it’s a collaborative team making the discovery, that’s just as good or even better! And sharing with others what you just learned has its own satisfying rewards. But, what if we left it there and failed to use that knowledge to improve nutrition, or increase sustainability of agriculture, or to realize benefits through some other important application? We would feel good growing our understanding of nature, but we would likely fall short of delivering on our mission.
I then talked about ways in which we enable Danforth Center discoveries, and the technologies we develop, to have tangible impact in the real world. It doesn’t happen magically; rather, it usually requires intention, investment, partnership and ultimately handover to companies that can build a better product, offer a better service, or otherwise deliver benefits in the marketplace. The Center’s Startup Initiative is designed to spin out companies that are based on Danforth Center discoveries. We get a “two-fer” if those companies succeed – our science gets put to work addressing grand challenges that we care about, and we create economic opportunity and growth in 39North and the region.
There are also other routes to impact, as I and Mike DeCamp (CEO of CoverCress, Inc.) described last night. For example, Meter Nusinow and his talented team have collaborated with CoverCress for many years to improve the light, temperature, shade and density response characteristics of pennycress, a recently domesticated cover crop. Discoveries from the Nusinow lab are directly transferable to breed pennycress that grows at higher latitudes, higher densities and with more resilience as growing temperatures increase. CoverCress not only benefits from the collaboration, but they benefit by renting plant growth and processing facilities from the Center.
The scientific process is one of humankind’s greatest innovations. We need it more than ever. We also need creative scientists, entrepreneurs, investors and mechanisms to transform that scientific knowledge into meaningful benefits, and that is just as satisfying as making a great discovery!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Toby Kellogg wrote a fascinating Perspective article in Science magazine recently. I look at all of Toby’s papers because they always address an interesting question in plant development or plant evolution (often both). In this case, Toby reviewed another fascinating article (Satterlee et al.) in the same issue of Science, where the authors asked: Why do some plants have sharp, pointy structures called prickles on their surfaces, but other plants in the same or different species do not? Prickles help plants defend again herbivores, who would rather move on to another plant than get poked by prickles.
Toby highlighted the key finding that, whenever a prickle-less mutant or natural variant was identified in different species, it was always because of a defect in a particular member of the LOG gene family. Nature through the process of evolution, as well as plant breeders through the process of domestication, came up with prickle-less plants independently on many, many occasions, and always by losing the function of the same LOG gene. That gene controls the production of an important plant hormone – cytokinin. Toby then pointed out that, besides answering an important question about how new traits evolve in nature, learning this type of information has practical benefit. Learning how a trait evolves in nature means we can apply that knowledge to breed plants quickly with that same trait, assuming that trait has value.
The prickles study reminded me of another study some 22 years ago by Lellis et al. using the model plant Arabidopsis, which is susceptible to a virus called Turnip mosaic virus (TuMV). Scientists created a large number of Arabidopsis mutants using DNA-damaging chemicals, and then tested thousands of individual mutants for those that lost susceptibility to TuMV. Losing susceptibility is another way of saying that the mutants became resistant to the virus. The loss-of-susceptibility mutants were found to have defects in a particular gene within the eIF-4E family, meaning that TuMV requires this host gene to infect Arabidopsis and cause disease. Once that basic information was learned, just like in the prickles study, scientists quickly figured out that crop plants like peppers and soybeans could be made resistant to TuMV-like viruses by breeding plants with similar defects in their eIF-4E genes.
Both the prickles and the TuMV studies are great examples of how plant science focused on a basic scientific question can lead to practical discoveries and applications. Many thanks to Toby Kellogg for crafting such a nice Perspective in Science, and for reminding me of those old findings with TuMV in Arabidopsis.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Let’s return to something mentioned in my message to the Danforth Center community on July 1, 2024. Referring to my intentions here over the next 11 months, I said “much attention will be focused on resource development for both current priorities and future opportunities, and on those things that promote success for the next President and the Center community.” Since that time, some of you have asked, what does that mean?
The short answer is, I’m focusing a lot of attention and effort towards strengthening those parts of the Danforth Center on which our current and future success depends. We’re a scientific research organization that requires competitive research teams and talented support units, enabling facilities in which to work, and routes to deliver benefits from scientific discoveries in the real world. Those are critical components of Center core strength, and key ingredients driving success of the institution. I’m focused on ensuring that our budget, fundraising and building efforts are aligned with these core priorities.
For example, considerable effort was invested recently to develop a plan to reprioritize $14.75M of existing Center funds, mostly from excess operating reserves and donor gifts, to strengthen core priorities. Half of these funds are focused on ensuring that we have sufficient resources to recruit and equip new Member-track faculty to fill open positions over the next several years. Most of the remaining funds are focused on ensuring that our research teams have sufficient capacity to make investments that maintain the competitiveness of their programs. I’m also working more closely with the Development team to ensure that core scientific and facilities priorities are supported as much as possible through successful fundraising efforts from donors over the next few years.
Bolstering core strengths that underlie success of the Danforth Center is a major objective of mine over the next year. The core priorities today will almost certainly be the core priorities tomorrow, meaning core investments today will pay dividends long into the future. These investments will be important for both current and future Danforth Center community members, including the next person to occupy my seat.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Did you know we run the largest Research Experience for Undergraduates (REU) program in the U.S.? This program brings in around 20 diverse students from around the country each Summer. It’s funded by a National Science Foundation grant, which is supplemented by funds from research grants to PIs and core Center funds. Today, I’m impressed and blown away by the 2024 students presenting their work in the 24th REU Symposium.
Year after year, each REU cohort exhibits interesting and sometimes surprising features. I spoke with several of you who were involved closely with the program to ask, what distinguishes this REU cohort? I was most pleased to hear the following.
- Collaborative and community-minded. This group bonded quickly with a Memorial Day BBQ just after arrival, initiating a cohesion that was evident throughout the program. They were often spotted in groups solving problems. They embraced, and integrated into, the broader Danforth Center community. The REU students were looking out for one another! And they formed a tight, entertaining karaoke team!
- Curious and insightful. This was an inquisitive and thoughtful cohort. On tours of core facilities or nearby companies, they were highly engaged and asked terrific questions. In the meeting with me, they were open, far-sighted, and intentional about developing their careers. They were very engaged and involved in every training session or outreach activity.
- Professional. Now, given that this may be our youngest REU cohort ever, this comment was so good hear. They were on time for all meetings, events and trips. When they took a trip to the Field Research Site, everyone was on time for a 7:25 am departure. When REU student Emmy Hood arrived in our lab, she was prepared though advance reading, accurate note-taking, and an orientation to get things done. A deaf REU student, MJ, even mentored some high school interns during the Summer! This cohort’s professionalism far exceeded their very early career status.
Many thanks to Kerri Peer and Monica Alsup (REU administrative coordination), Tessa Burch Smith and Kirk Czymmek (REU Program Directors), and Katie Murphy (Phenocore Director) for your comments about the 2024 students and your great work with the program. My sincere appreciation goes to the lab mentors who guided each REU participant through their projects. And congratulations to all of the 2024 REU students for what they achieved for their careers and contributed to the Danforth Center. I hope to see them back in the future.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Feedback is necessary, useful, and often illuminating. Feedback leads to self-reflection, which is a step on the way to improvement. I hope everyone takes advantage of the upcoming Engagement Survey Listening Sessions (starting July 15) to offer feedback about your experience at the Danforth Center. I’ve received or heard about a lot of feedback from individuals who recently visited the Center, mostly as candidates interviewing for new Assistant Member faculty positions. As their feedback is largely a reflection on the Danforth Center community, I think you’ll be interested in what they said.
“This is the 8th Wonder of the World.” This direct quote was from a scientific collaborator at another institution. Our site, our facilities, and the way our facilities are maintained are noticed and admired by visitors. Our facilities are exceptional and often envied.
“Wow, you seem really collaborative.” The five recent Assistant Member candidates all commented on this. They were particularly impressed by how nearly all projects are collaborations, and how scientific teams have evolved over time based on those collaborations. The candidates were encouraged to envision how their programs would impact other Danforth Center teams through collaboration.
“The scientific culture seems really different here.” The recent candidates noticed a culture of sharing resources and sharing credit. They commented on how our mission, vision and values really shape what we do and how we talk about our work. They were impressed by a culture of getting things done, and how administrative and non-research community members contributed to and shared ownership in scientific success. They were surprised how efficiently the search was done and by everyone’s helpful attitude.
“The trainees and CSTM members were really engaged.” Each candidate commented about how impressed they were with CSTM members’ participation in the search. They noticed a sophisticated understanding about both the needs as well as the opportunities for mentoring, and at least some of the candidates found this to be eye-opening.
“I’m on board!” This was from one of the candidates, Nadia Shakoor, who accepted our offer of Assistant Member just this morning! We’re excited for Nadia and her team, and for the new opportunities for growth at the Danforth Center. I hope you join me in congratulating Nadia on her new position and for all that she’s done.
Jim Carrington,
President and Chief Executive Officer
Note: The Weekly Message will be on hiatus for a few weeks until I return from vacation. I’ll be out to pasture, think it’s safe to say, enjoying the Rocky Mountain way just south of Mt. Sneffels.
Dear Danforth Center Community,
I hope everyone had a chance to relax or enjoy festivities during the 4th of July holiday yesterday. Those who were concerned about outdoor events getting rained out may have spent some time watching televised sports, like the early rounds of Wimbledon tennis on ESPN. Had you stayed tuned after tennis, ESPN broadcasted the Annual Hot Dog Eating Contest from Coney Island, NY with a full complement of sports announcers and analysts. Now, I do not consider myself strongly anti-hot dog, especially with the grandson of the inventor of the Ballpark Frank being a Danforth Center community member. But I have a big problem with classifying the over-consumption of hot dogs, or any processed meat for that matter, as a sport. C’mon, man!
Had you kept ESPN on in the background as you prepared to view night-time fireworks, you may have seen the Annual Diving Dogs competition, featuring some true canine athletes. I can only describe this as a hybrid of long jump and competitive fetch, but into a swimming pool. In the featured event and with a jump of 28 feet, a Whippet named Rogue edged out the favorite, Sounders, the one everybody came to see and the holder of eight Diving Dogs world records. I’ve never seen athletes enjoy competing as much as these dogs.
However you spent the day off, you probably did yourself some good. Besides increasing productivity and overall life satisfaction, taking time off from work leads to both physical and mental health benefits, including lower rates of heart disease, reduced depression, lower stress levels, and better sleep. Time off work also gives you a chance to have fun, and fun is highly underrated!
Finally, I want highlight those who deferred time off or worked unusual hours on or around the holiday. Many of you were needed for plant care, security, essential lab work, or unanticipated emergencies on July 4th. Others, like the team that handles payroll, rearranged schedules to ensure work was completed earlier than usual. I give sincere thanks to all those who kept the Danforth Center running through the holiday. They deserve our appreciation!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
When I came to the Danforth Center as President in early 2011, there was no thought about how long I’d be here. There were too many possibilities at hand, and so much to build and achieve together with the Danforth Center community, our partners and our supporters. But over 13 years later, the time has arrived to ask the question, “How long?” After much consideration, I’ve decided to step down as President and CEO of the Danforth Center after one more year, on July 1, 2025. This decision was made with the Danforth Center’s future, my family, and sexagenarian biology in mind, and with peace in feeling that it’s time.
Planning for succession and identifying the next leader, like everything at the Danforth Center, will be a collaborative effort. Penny Pennington (Chair, Board of Directors) will lead the search for a new President. Penny will appoint a search advisory committee, which will include representation from the Board as well as the Danforth Center community. She will start communication soon about the search process, the anticipated timeline, and how input from the Center community and stakeholders will be received.
In addition to my current roles, responsibilities and goals over the next year (I’m not done yet!), I’ll be working to ensure that opportunities for the next President, and for members of the Danforth Center community, are as abundant as they were for me when I started. That means much attention will be focused on resource development for both current priorities and future opportunities, and on those things that promote success for the next President and the Center community.
I love the Danforth Center, our people and our shared purpose. I expect that I’ll always be a part of this unique and impactful institution. And I know the Center will benefit greatly from the necessary renewal of perspective, ideas, and talents that come with a new leader. In the meantime, I look forward to one more year of achievement with you in this position I’ve been so fortunate to hold.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Pat Brown - founder of Impossible Foods, Director on our Board, and recent answer to a clue on Jeopardy! - is one of the most influential biologists over the past 40 years. And with a goal to eliminate animal meat production on a global scale, he’s also one of the most influential food innovators over the past 15 years. Pat was invited recently to a Royal Society meeting on transformation of agriculture, at which he described himself as the designated provocateur! A few of us connected with Pat by Zoom this week to seek his input on a few matters and to give some Danforth Center updates. Here’s a little of what he had to say.
We can grow enough protein for human consumption in plants. Pat is a champion for reducing the environmental impact of agriculture through reducing/eliminating the use of animal meat in our diets. A few simple calculations indicate that we could provide more than enough protein directly from plant-based agriculture. Human consumption of protein from meat worldwide currently stands at about 90 million metric tonnes (mt), while protein produced by soybean cultivation alone amounts to nearly 170M mt. Of course, there would be enormous challenges - dietary, cultural, economic, and technical – to seeing that substitution happen. Concerning the technical challenges…
We need more protein innovation. Realizing Pat’s vision requires that we develop more nutritionally complete, digestible, accessible and safe plant-based protein sources. He sees a wide-open opportunity to apply science to produce at scale proteins that have complete amino acid compositions, better functional properties for food preparation, and greater production efficiency with fewer inputs. He ponders, might we be able to use vegetative plant parts (e.g. leaves), rather than primarily seeds, to produce next generations of plant-produced protein?
Need better understanding of everything underground. Pat is a big proponent of not only reducing animal agriculture, but also of diversifying our agricultural crops and gaining a better understanding of what’s happening below ground. Carbon capture, reduction of greenhouse gas emissions, and other benefits will be gained by placing far more attention on roots, soils, and subterranean environments. Pat, you’re speaking our language!
It's safe to say, not everyone agrees that Pat’s objective of eliminating animal agriculture is practical or right. But it’s hard to argue that he not driving innovation, behaviors, and markets that are providing measurable environmental benefits.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Let’s start with a quiz. What’s one of the biggest differences between someone who gets a meaningful award and all of their worthy peers who did not? Answer: The award winner had a nominator!
I want to make the strong case for each of us to recognize and celebrate our high-achieving peers, supervisors and direct reports with nominations for awards and honors. This requires someone, a nominator, to spend time and effort assembling a packet of information that describes why an individual has earned recognition. Investing valuable time to submit a nomination is a selfless act of commitment to, and appreciation of, a colleague. It is a high form of respect for what they’ve done. Preparing a nomination means you care enough to advocate formally for a colleague who did well. And when you’re on the receiving end, awards can be powerful affirmation that you’ve made an impact through achievement.
Awards and honors come in all shapes and sizes. There are scientific society awards that recognize a variety of research, scholarly, educational and other contributions. There are awards for individuals in all professional disciplines, like finance or human resources. At the Danforth Center, we give awards to high-achieving graduate students and individuals who have made major contributions to plant science research and service, and we are looking at ways to recognize outstanding members across the Danforth Center with new annual awards. Nominators are the keys that unlock awards for our deserving colleagues.
Many of you are active nominators, and in so doing you are making a major contribution to the Danforth Center community. Thank you! But I ask that everyone spend a few moments and ask, “Which of my Danforth Center colleagues are deserving of special recognition?” Whether it’s a major ASPB award for career-long achievement or recognition as CSTM Member of the Month, being recognized matters. And if you don’t do the work to nominate a coworker, then who will?
Congratulations to all of our recent award winners, like Karla Roeber, who is being recognized this week as Outstanding Member of the Year by the St. Louis Agribusiness Club. Well done, Karla, and many thanks to your nominator!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
One of the organizers said the best thing about the annual Danforth Center Scientific Retreat is the way scientists and teams come together and learn what everyone is doing. The 2024 Retreat this week welcomed over 200 participants, including Penny Pennington (Board Chair) and Silvia Restrepo, President of our peer institution, Boyce Thompson Institute. I’ve been impressed by the quality of every short and long presentation, and the progress shown by each speaker and team. Here, I want to highlight a few presenters who also made their work especially accessible to a broad audience.
Keely Brown talked about developing rapid, predictive breeding methods applicable to perennial crop improvement. Perennial crops present unique challenges, like the multi-year nature of cultivation and long life cycles. Keely is using data science, genomics and phenotyping approaches to more quickly breed perennial crops with high yields and environmental benefits (e.g. high carbon sequestration) in different geographies.
Dhiraj Srivastava is working collaboratively within the Phenotyping core to devise better ways to capture and analyze data from field experiments. He is using drone-based sensors and methods, for example, to measure height of both short and tall varieties of tef over time. He’s extending the PlantCV platform to include important geospatial capabilities. Dhiraj is a great example of a scientist who organizes presentations around a key principle: Make it understandable to a broad audience!
Kris Callis-Duehl gave an overview of the Education Research and Outreach Laboratory (EROL), which seeks to combine science and education to achieve STEM+Ag objectives for students and teachers, workforce development, understanding of how students learn, and more. EROL is a unique program that has impact on economic development, especially in underserved communities in our region, as Precious Hardy elaborated on very nicely in her short talk. The EROL team also works closely with many Danforth Center lab teams for broader impacts through outreach, and for data science skills training as Parag Bhatt described well in tie-dyed color.
I also noticed that Dan Lin, Dominique Pham, Stewart Morley, and Cody Bagnall included notable features in their presentations to make their complicated topics understandable or even entertaining. Learning what our Danforth Center colleagues are doing promotes unity within the community and a strong scientific culture. The time and effort each presenter invested to bring their work to life for all is sincerely appreciated!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Change is a constant at the Danforth Center. It describes our scientific research programs, our people and our facilities. Change is necessary to grow, important for competitiveness, and essential to maintain relevance and impact. Let’s highlight a few changes and celebrate those working hard to make them happen.
Summer REU Program. The Center changes at the end of May each year with the arrival of 20 or so interns in the Research Experiences for Undergraduate program. The summer program has changed much over time, including this year with a younger cohort in which most have just finished just their first or second college years. They will be engaging in a new outreach activity at Oak Bend Branch of St. Louis County Library, where they’ll help 7-14-year olds learn to extract strawberry DNA. I met with the REU students this week and was exceptionally impressed. I sincerely thank them for investing their summer with us.
New Faculty Hiring. Prospects for significant changes within the faculty ranks improved dramatically with a new funding plan to hire for six positions over the next several years. We’re off to a fast start! Over the next six working days, we will host six candidates interviewing for two Member-track positions. New faculty hiring means new teams, changes in our capabilities and capacity, and renewed energy to power unique collaborations. I thank everyone with a hand in organizing the searches, reviewing applications, meeting with candidates, and otherwise participating to show off the best of the Danforth Center.
New working spaces. Everyone can see the new office and meeting facility under construction next to the greenhouse complex. But there are out-of-view changes underway to create new spaces for Grant Specialists and (soon) the Finance team. The Facilities team is simply the best I’ve seen at addressing the ever-changing needs of an organization like ours. Chris Martin, for example, is now renovating one of the 3rd-level office suites to grow capacity. Come to think of it, Chris always seems to be working hard on a building project, repairing something, or changing a space to accommodate different users. Chris and the rest of the Facilities team deserve our sincere appreciation for constantly improving and evolving our facilities.
Oh, I almost forgot one more change…Karaoke is coming back to the Scientific Retreat next week. But this time, all Danforth Center community members, not just retreat participants, are invited! I look forward to hearing your collaborative efforts at the microphone.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Let’s build this week’s message around two colorful images that I’m excited to share. Both represent important teams or features that we value at the Danforth Center, and that make us unique.
Grant Specialists, Administrative Assistants, and our scientific network. Each year we build a network diagram, thanks to Noah Fahlgren, Melissa Kerckhoff, and Karla Elliott, showing how our scientific teams connect through funded research projects. It’s a representation that dramatically underscores our value of Collaboration within the Center’s scientific enterprise. Not surprisingly, Grant Specialists and Administrative Assistants have some of the most connectivity within the network. Collectively, these two groups facilitate accurate grant proposal preparation and on-time submission. In 2023, their work resulted in 82 submissions and $23M in new grant awards. They help build and manage budgets, and bridge between collaborating teams. They help plan, organize, and run seminar programs, retreats, internship and education programs, and so much more. And they each work across multiple scientific or core facility teams, and frequently with the pressure of tight deadlines. They are critical hubs that hold the network together! With admiration for the vital work they do, I thank the Grants team of Michelle Richards, Missy Rung-Blue, Alex Durdello, Denise Cunningham, Elizabeth Martinez, and Marti Meersman; and the Administrative team of Terri Burton, Monica Alsup, Shannon Gabbert, Kerri Peer, Judy Mitchell and Jenny Kezele.

New art is coming! Last year, I communicated about feedback from the Danforth Center community concerning the artwork displayed on the 2nd level. That feedback led to formation of a committee, composed of Kerri Gilbert, Sarah Jennings and Anna Dibble to develop ideas for new art that would better reflect our community and purpose. After much investigation, consultation, and consideration, I’m happy to announce that we will install two colorful “moss walls,” composed of preserved mosses and other plant materials, in place of the current four paintings. The Danforth Center prairie was the inspiration for the design. Look for the installation to begin within the next few months. Many thanks to Kerri, Sarah and Anna for your creativity, time and effort.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I spent part of yesterday with a group discussing the following difficult questions: What can we do to better educate and inform “the public” about science and evidence-based decision making, and what can we do about rising anti-science influences in the U.S.? It was helpful that Dr. Sudip Parikh [CEO of the American Association for the Advancement of Science (AAAS)], and Lauren Seligman (AAAS Senior Director) were part of the conversation. With a mission to “advance science, engineering, and innovation throughout the world for the benefit of all,” AAAS is one of the most important organizations supporting, promoting and advocating for science and research.
The reasons people adhere to beliefs that have apparent anti-science foundations are often not because they are generally anti-science. Among other reasons, social, political or other group affiliation and acceptance, and the information that groups share among members, has been shown to effectively drive and reinforce beliefs independent of consideration of evidence for and against the beliefs. But the same person who argues, for example, against evidence that people set foot on the moon may be a staunch defender of the science and scientists behind a modern anti-cancer treatment that saved their life. The difference is this: the life-saving medication mattered in a most personal, compelling way; belief that the moon landings were a hoax is an option with no apparent or tangible consequences.
The AAAS under Dr. Parikh’s leadership is taking a strategic approach to infuse better understanding of, and comfort with, science and evidence-based decision making within society. In addition to all of the work with scientists, scientific organizations, and educators, AAAS works closely with specific, influential groups to provide targeted education, resources and support. These groups include legislators and elected officials, federal judges, and religious leaders. Working with the latter starts with acknowledging that both faith-based beliefs and science in our lives can and must productively coexist. And the AAAS Science and Technology Policy Fellowship program embeds 250 talented scientists and engineers across all three branches of the federal government, infusing scientific and evidence-based perspectives in offices and agencies that need them. The AAAS approach is based on non-partisanship, relationship-building and understanding of the challenges we all face.
I thank Dr. Parikh and Lauren for the great work they’re doing with AAAS for the benefit of all, regardless of ideology, group affiliation, or political identity. I’ve been a card-carrying member of AAAS for most of my life, and now you know some big reasons why.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
When we reported productivity metrics to the Board of Directors last week, I was thrilled to comment on the 40 graduate students who did dissertation research at the Danforth Center in 2023. These students are highly valued contributors, collaborators and colleagues, and we would be a far less interesting, dynamic and fun place without them. Yesterday, I had the opportunity to spend time with four Washington University (WU) Ph.D. students, including Paty Gallardo (Bart team) and Sarah Pardi (Nusinow team) here at the Center, several WU faculty, and Vi Skukla (Senior Program Officer, Bill and Melinda Gates Foundation.)
Much of the far-ranging conversation dealt with how scientific research and the lab environment has changed for graduate students over the years, including during the pandemic. We talked about how the pandemic affected both the preparation for graduate school and life as a student, and what might be the long-term consequences of those difficult years. Paty felt that she learned much less as an undergraduate student during the pandemic and entered WU with too little preparation compared to her peers. (Editorial note: Paty is one of the best prepared graduate students we’ve ever seen!). As a graduate student beginning her research when the pandemic hit, Sarah had to manage all of the rigorous program requirements in addition to science at the bench. Sarah and Paty have excelled despite the headwinds.
We also talked about how remote learning, absence of in-person scientific meetings and overall less direct human interaction during pandemic years might affect career development beyond graduate school. Will students who traversed the last four years be able to build robust professional networks, which are vital to navigate all parts of the scientific research world? I have serious concerns about losses and setbacks of talented individuals from research career paths over the last several years. But if you’re looking for examples of resilient scientists who inspire confidence about future research leaders, spend a little time with Paty and Sarah.
Now, while I was writing this message, Sarah Pardi participated in graduation ceremonies to recognize the award of a Ph.D. Congratulations, Dr. Pardi! It’s a world-class understatement to say you earned it.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
In late 2022, I updated the Danforth Center community on progress towards sustainability initiatives in our facilities, highlighting energy conservation measures, use of renewable energy, and progress toward our goal to reduce greenhouse gas emissions to 28% below 2005 levels by 2025. I also highlighted a few areas in which we needed to ramp up our efforts, like reduction of single-use disposable plastic. Since then, we obtained a sustainability audit by an independent firm, which helped with both short and long term planning. Let’s discuss recent progress and upcoming actions that will move us forward on three fronts.
Reduction of lab plastic waste. We recently installed a Grenova TipNovus Tipwasher in the lower-level media kitchen for cleaning and reuse of plastic pipette tips. The system can complete a wash and dry cycle for four boxes of commonly used, non-filter pipette tips in 25-30 minutes. Although there are limitations with certain tip sizes, all labs are strongly encouraged to explore and use the system to reduce a significant source of plastic waste. I am grateful to Becca Bindbeutel, who will train lab members on use of the tipwasher. Becca is also exploring ways to reduce or eliminate non-recyclable, disposable lab gloves.
Reduction of café plastic waste. The café, which is now part of the Green Dining Alliance, has completed the transition to compostable or recyclable materials for all take-out packaging, containers, bowls and utensils. The recycle and compost bins by the café are now larger and easier to use, and will soon have improved signage to guide use. Many thanks to the café team!

Solar energy at the Danforth Center. Say goodbye to the tents, and say hello to a new solar power-generating pavilion that will be constructed later this year on the Miller Terrace! The solar panel-covered structure will yield an amount of electricity to offset twice that needed for the new Plant Growth Facility office building under construction, and reduce 78-109 metric tons of CO2 emissions annually from fossil fuel-powered electricity generation. Also, through Ameren’s Renewable Solutions Program and the purchase of Renewable Energy Credits, we are helping bring online and use a new solar facility in the region. We anticipate spending up to $50,000/year on this program, and meeting our greenhouse gas reduction goals by the end of 2025.
Shrinking our environmental footprint is a continuous process, and I thank everyone for doing their part.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
The first thing Penny Pennington, the fourth Chair of our Board of Directors, brought up in a meeting with 35 or so Danforth Center community members earlier this week was the birth of her first grandchild, Quinn, just a few days earlier. The rest of her conversation with the group, including why Penny chooses to work with us in addition to her day job as Managing Partner (CEO) of Edward Jones, was also inspiring. In Penny’s own words, I want to share portions of that message with the entire community.
“My ‘why’ is all about helping more people thrive…I want to invest in organizations and in people who are making a difference in the world outside of Edward Jones. I just want to be part of what you do. I want to help facilitate, I want to help enable you and the Danforth Center to do what you do. When I became Managing Partner in 2019, the first off-site that I did with my senior leadership team was here…I wanted us to have an experience of being in the environment that you create. And it's an environment of great ambition. It's an environment of having a North Star that really matters.”
It's an environment of innovation, of collaboration, of looking long-term and saying, ‘what can we do to make a difference in the world using the skills and the tools and the expertise that we have?’ So that's my ‘why’ for Danforth Center (engagement) and my thanks to you for what you do. I want to be the best version of a Board Chair that I can be. I want to learn from you…I'd really like to spend some time with you.
I've now got the fourth generation of my family alive. And I tell you what, there's nothing like having another generation to say, my gosh, what is happening in the world and what can we do to make a difference that really matters for people. It's never mattered more to me than it did as of a week ago when Quinn was born.”
Penny’s devotion of time and effort here is an investment in each Danforth Center community member. If you see her in the hallway, please introduce yourself, ask how Quinn is doing, and let her know what you do. I’m certain she will appreciate learning how her investment is growing.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
If you’re a fan of those who know what they need, and then roll up their sleeves and get to work, you will be impressed with the Committee for Scientific Mentoring and Training (CSTM) at the Danforth Center. As a voluntary assembly of postdoctoral trainees, graduate students, research associates, technicians and other non-faculty scientific professionals, CSTM members seek to acquire new skills, enhance their careers, build community and make life better at the Center. The group is led this year by co-chairs Vanessica Jawahir, Melette DeVore, Myia Elliott and Dhiraj Srivastava, with some notable big lifts by Seth Edwards and Lily O’Conner, and they have a significant agenda. Besides the annual BioBash and PlantTech Jam events, here are just a few of their priorities for 2024.
Science communication skills. CSTM recognizes the high value of being able to engage scientific and non-scientific audiences better, so a workshop focusing on public communication is being planned. They want to learn the skills of those who connect with and inspire broad audiences so well. I applaud this and plan to participate by sharing a thought or two.
More social events. Wait, is this just about having fun? Well, CSTM members understand that fun is highly underrated! They also understand that a work environment in which you know, appreciate and enjoy spending time with your colleagues is a more productive and satisfying workplace. Social events like monthly happy hours and casual gatherings are also golden opportunities to stir up new ways of thinking about research. As Vanessica told me, “We come up with more innovative projects when we interact with people who have diverse ideas.” It’s the “interact” part that is critical, and it’s vital for the culture we desire.
Recognition of CSTM members. The team is creating more opportunities to recognize and celebrate their peers. Did you see that Somnath Koley, Research Scientist, was the March 2024 CSTM Member of the Month? This kind of recognition is important not only to call out those making significant scientific contributions at the Center, but also to learn more about our diverse community. The co-chairs highly encourage new nominations! Also, Travel Awards recognize and help those CSTM members who are gaining professional exposure at scientific conferences.
The secret sauce of CSTM is high motivation and willingness to explore new routes to career growth, to build a supportive culture, and to work hard. I’m a big fan of CSTM!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
We recently held a one-day Faculty Retreat at the Forest Park visitor center. I always look forward to this annual event. It’s an investment of time and effort to concentrate on issues that directly or indirectly affect every team at the Danforth Center. It is particularly useful to gauge opinions and gain input on potential investments, resource allocation, new opportunities and issues of concern. So, what did we cover?
Space Allocation. We reviewed and discussed the near-final draft of a new space allocation policy, which will be available soon on Workvivo. As communicated in the January 19, 2024 message, space allocation is challenging because space is a limited resource. We discussed the key factors when assigning space – What are the needs, what is available, and what is reasonable within the broader context (e.g., competing needs or future plans). We also discussed the goal of achieving fairness, in part through transparency.
New Faculty Hiring. Encouraging progress was reported on searches for a new Assistant Member and a mid-career Mizzou-Danforth Center joint faculty member. I am optimistic we’ll have two new research team leaders later this year.
Sources of Funding and Funding Needs. Sources of Danforth Center funding for projects, equipment, graduate students, and other research needs were reviewed, followed by a poll and lively discussion of pressing needs for flexible research funding. I have committed to work with Becky Bart (Interim VP for Research), and Hal Davies and Djuan Coleman (Finance), to review options and identify funding sources to address some of these needs within 2024.
Field Phenotyping Opportunities. A significant grant was awarded to both refurbish the Bellwether Phenotyping facility and develop new phenotyping capabilities at the Danforth Center Field Research Site. We have the opportunity to build or adapt technologies to elevate our programs. The discussion and workshopping focused on possible forms and functions of new technologies or facilities that would meet the needs of the most Danforth Center teams. The wisdom of the retreat crowd will be incorporated into plans under development now.
Developing Leaders. We reviewed and discussed the range of leadership training opportunities at the Center, and heard from several recent participants in the Developing Scientific Leaders program. I am sincerely thankful to all who using these opportunities to become better team leaders.
I also appreciate everyone’s contributions to an interesting and productive Faculty Retreat!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
“Simplicity doesn’t need to be greased.” – Billy Joe Shaver
I would bet that most of you have never heard of Billy Joe Shaver. He was a highly respected songwriter and a founding force of the outlaw country music movement in the early 1970s. He wrote dozens of notable songs for Waylon Jennings, Willie Nelson and many others. Though he was hardly a role model in life, he provided this absolute pearl of wisdom. Though his statement about simplicity dealt with the songs he wrote, I believe it can be applied by anyone wanting to achieve anything important. Here’s why.
Simplicity helps focus on the most important goals. Simplicity is not about aiming lower, but rather, helping to focus on the most important work to achieve goals. I remember serving on a review board for a scientific institute in Europe. After questioning one of the PIs about their very long, complicated report, their response was a semi-serious, “Yes, my team is tightly focused on 16 different projects.” How many of those projects were distracting? Were the most promising projects under-resourced? Were team members receiving the attention they needed?
Simplicity is more efficient. In terms of achieving a big goal, I always advise to concentrate really hard on the next step, as opposed to over-worrying about the next six steps. That does not mean ignoring or abandoning a plan, but rather, keeping attention focused on achieving meaningful progress. There is no substitute for progress! Distraction due to over-complication is exhausting, expensive, progress-inhibiting and demoralizing. Teams that are able to simplify steps move faster, and are able to anticipate, understand and respond to problems more efficiently.
Simplicity is easier to understand. This is a big one. How many scientific seminars or technical training sessions have you attended where the speaker used excessive acronyms, complicated and specialized language, or an overbearing Powerpoint presentation? Even those who are specialists in the particular field are often confused due to the complexity and coded language. Simplifying a message is a gift to everyone listening to you talk or reading your report. Those who are unfamiliar with the topic have the possibility to learn something important or be inspired, and the specialists in the field will appreciate it because it’s refreshingly different!
Might you or your team benefit by adopting more simplicity? I’d love to hear your thoughts.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
You would never confuse Phil Needleman for a card-carrying plant scientist. Rather, Phil was a rigorous biochemist and pharmacologist who made life-changing discoveries that impacted millions of people. Phil discovered key parts of biochemical pathways that influence blood pressure, blood clotting and inflammation. He led teams that studied the enzyme COX-2 and COX-2 inhibitors, leading directly to the anti-inflammatory drug, Celebrex, for the treatment of chronic pain associated with arthritis and other conditions. He was highly recognized, awarded and admired for his vast contributions to science and medicine.
Despite his lack of plant science bona fides, Phil made tremendous contributions to the Danforth Center. In 2005, he was cajoled by his friend Bill Danforth to join our board of directors, where he remained for nearly 19 years. He was the Center’s interim president for over a year, providing both leadership and stability prior to hiring of new guy in 2011. From my perspective, Phil made his most important contributions in three ways. First, he was a scientist’s scientist. He cared deeply about the importance of a scientific question, the quality of an experiment, and the clarity of results. Phil believed in data. He wanted to see it, and he wanted to talk about it…a lot! He once had a sign on his door that read, “If you have data, knock then enter. All else requires an appointment.”
Second, Phil constantly encouraged, demanded really, that we apply what we learn to solve big problems in the world, and he supported us in doing so. In part because he was inspired by many of you, Phil became interested in the Nitrogen Problem in agriculture – there’s too much greenhouse gas emission and other environmental impact due to current reliance on synthetic nitrogen fertilizers. He wanted to learn what we were doing about it, but he was even more interested to work with a team to organize new approaches. This was how the Subterranean Influences on Nitrogen and Carbon (SINC) center was conceptualized, and then launched through generous donations from Phil and his wife, Sima.
And third, Phil provided wisdom, mentoring and personal support to so many of us at the Center and on the board. At this point, I have a confession to make. On a few occasions, I plotted covertly with Phil ahead of a board of directors meeting. Before proposing and seeking approval for new ways in which to create start-up companies, for example, we had this exchange. “Phil, I know you support this, but I need you to help make the case in the meeting,” I said. “Don’t worry," Phil replied with his usual wide grin. "I’ll be ready with fist-pounding support when the time comes." Phil relished opportunities to help me and countless others.
Phil Needleman’s life ended abruptly this week. But his legacy, example and will to make the world better through science lives on.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Several of us hosted a visit today from the Honorable Francis Gatare, cabinet-level head of the Rwanda Development Board. As leader of the agency that oversees the national economy of Rwanda, Francis Gatare has a mandate to accelerate and enable Rwanda’s economic development through private sector growth. You might be surprised to learn that Rwanda is the most densely populated country in Africa. Ninety percent of employment comes from the food and agricultural sector, which accounts for 35% of Rwanda’s gross domestic product.
From his position leading Rwanda’s economic growth, Francis Gatare’s top priority for agricultural economic development is increasing productivity of crops. And that’s where the Danforth Center is playing a role. In 2019, colleagues at the Rwanda Agricultural and Animal Resources Development Board (RAB, the national agricultural research organization) joined Virus Resistant Cassava for Africa (VIRCA), the Nigel Taylor-led effort funded by the Bill & Melinda Gates Foundation and USAID to address the major virus diseases of cassava in East Africa. Given that earnings from cassava exports declined by 40% due to Cassava brown streak virus between 2016 and 2017 in Rwanda, there’s a great need for resistant varieties. The VIRCA goals in Rwanda include testing performance of recently developed varieties in field trials, training scientists, obtaining approvals, registering improved varieties, delivering seed to smallholder farmers, and training entrepreneurs.
“Wait, what’s with training entrepreneurs,” you ask? One of the biggest challenges to delivering virus-resistant cassava in target geographies, like Rwanda, is the lack of effective, sustainable seed distribution systems. There are no companies to scale-up planting materials, steward varieties, and sell improved cassava seed to farmers. To reach 90% of the 700,000 cassava farmers in Rwanda, the VIRCA team aims to help develop and train 250 Cassava Seed Entrepreneurs (CSEs) who would multiply, quality-assure, and sell cassava seed to farmers. The CSEs will build private businesses that serve an unmet need and provide a source of income. In partnership with Rwandan entrepreneurs, developing private-sector cassava seed systems that are economically sustainable is now one of the top objectives of VIRCA.
I was thrilled to meet Francis Gatare and discuss the opportunities to both increase agricultural productivity and build economic strength in Rwanda through a partnership about which we should all be proud.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Today, I’m honored to celebrate International Women’s Day and recognize the remarkable community of women at the Danforth Center. This year’s theme is Inspire Inclusion, which emphasizes the importance of diversity and empowerment of women in all aspects of society. Inspire Inclusion is about encouraging everyone to recognize the unique perspectives and contributions of women from all walks of life, including those from marginalized communities.
I’m inspired every day by women at the Center and the unique contributions they make. Within the teams in which I work and collaborate, I appreciate Allison Brown’s and Diane Moleski’s commitment to our community through their organizational skills. I am thankful for Kira Veley’s determination and Kerri Gilbert’s creativity. I admire Myia Elliott’s desire to grow professionally and help others. And I celebrate collaborator Becky Bart’s inventiveness and abilities to reach lofty goals. Across the Center, I know each of you are similarly inspired by women on your team or those you work with regularly.
In addition to International Women’s Day, March 8 this year is also Grant Professionals Day. Talented contributions from proposal preparers, managers, and grants finance team members, nearly all of whom are women, are absolutely essential for our work and our success. The Grant Specialist team members – Kelly Brinton, Alex Durdello, Denise Cunningham, Elizabeth Martinez, Marti Meersman, Missy Rung-Blue and Michelle Richards (Grant Support Manager) – are sincerely appreciated for their professionalism, responsiveness, tenacity, and endurance. I have comparable admiration for Finance team members - Cathy Kromer, Melissa Kerckhoff, Jackie Gonzalez, Heather Bowen, Darine Kube and Julia Ruvinov - who have key roles in proposal submission, grant fund management and compliance. I also point with high praise toward Development team members – Lee’at Bachar, Sarah Jennings, Matt Wichmer, and Debbie Davis – who collaboratively organize, write and manage philanthropic proposals and subsequent gift funds.
I hope everyone takes some time to honor and appreciate the inspiring women at the Danforth Center, as well as the committed community members who make grant proposals and grant funding possible. Today reminds me just how fortunate we are!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
One reason I periodically highlight a few research articles published by Danforth Center scientists is because it helps me dive deeper into what our teams are discovering and thinking about. Here are a few such articles that I enjoyed reading this week.
Murphy, K.M., Ludwig, E., Gutierrez, J., Gehan, M.A. (2024). Deep learning in image-based phenotyping. Annu. Rev. of Plant Biol. doi: 10.1146/annurev-arplant-070523-042828.
This expansive review focuses on deep learning, a form of artificial intelligence using deep neural networks, to extract meaning from the fast amount of image data collected through phenotyping research. Teams here and elsewhere face unprecedented bottlenecks when analyzing massive datasets, and AI-based approaches are the only practical options to gain insights at scale. Katie and co-authors provide a great review of the progress, promise and challenges with deep learning for plant scientists.
Li, M., Liu, Z., Jiang, N., Laws, B., Tiskevich, C., Moose, S.P., Topp, C.N. (2024). Topological data analysis expands the genotype to phenotype map for 3D maize root system architecture. Frontiers Plant Sci. doi: 10.3389/fpls.2023.1260005.
Speaking of phenotyping, Mao Li and colleagues analyzed the genetic underpinnings of phenotypic variation of three-dimensional root characteristics between maize lines that differ in their nitrogen uptake and assimilation properties. This team is pushing the boundaries for how to use image-based phenotyping, mathematics and statistics to learn how plant roots take their shape. They demonstrate that the genetic basis for root architecture, and also the ways to describe and quantify root architecture, and far more complex than we currently understand.
Sankoh, A.F., Adjei, J., Roberts, D., Burch-Smith, T.M. (2024). Comparing methods for detection and quantification of plasmodesmal callose in Nicotiana benthamiana leaves. Molec. Plant Microbe Interact. doi: 10.1094/MPMI-09-23-0152-SC.
Amie Sankoh and the Burch-Smith team are pioneers in discovering how plants transmit information, important molecules and invasive viruses between cells. For example, they are leading the way in showing how plasmodesmata (PD) – channels that connect adjacent cells in leaves and other organs – are controlled by deposition of callose (a carbohydrate polymer) when plants are responding to infection. This article compares different methods to rigorously quantify callose accumulation around the microscopic PD channels, and paves the way for deeper understanding of how the plant immune system works.
Congratulations to each co-author on these published studies. Well done!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Let’s start with a statement about a primary function of the Danforth Center: We are in the business of creating new knowledge. We’re a research organization built firmly around the idea that learning something new can lead to something better in the world. Scientific discovery through our research is an engine that powers innovation, new technologies, new applications, and new or better businesses that address big challenges in the world.
Scientific research is hard! For organizations like ours, coming up with topics to study and ideas to test require a unique combination of deep knowledge, imagination and creativity. Grant proposals to fund research projects are time-consuming and exhausting, and only a minority of those get funded. Designing and executing experiments is tedious, laborious, and subject to failure and frequent restarts. There are no guarantees that productive, publishable advances will occur. But far more often than not, scientists will tell you, “I love it!” Why is that?
There are countless answers I could give, or that other scientists have told me. Research offers a feeling of purpose, and that’s part of what unifies us at the Danforth Center. One colleague recently told me that contributing and collaborating with other researchers, and seeing ideas and results validated, is really fulfilling. For me, the discovery process to explain a part of nature is like putting together a puzzle, except you don’t know how many pieces you have and there’s no picture on the box! It’s a challenge that rewards curiosity and persistence.
But there’s something else about discovery that’s both deeply personal and magnificently communal. When you make a discovery, or for the first time understand the answer to a previously unanswered question, you possess a piece of knowledge about the natural world that nobody else knows! It belongs exclusively to you or a small handful of team members. The reveal to your colleagues and the rest of your scientific community is even better, as is the satisfaction of being recognized for you’ve done.
I believe the culture of discovery should encompass all corners of the Center, not just the PI-led research teams. Might there be opportunities to better integrate elements of scientific discovery into, say, philanthropic fundraising efforts or development of Center facilities? I’m proud of the culture of discovery at the Danforth Center and believe it should be nurtured and grown.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
With recent or upcoming retirements that were announced recently, and the upcoming departure of Blake Meyers and part of his team, I’ve been thinking a lot lately about comings and goings. Research organizations like the Danforth Center experience a steady flow of individuals leaving for other opportunities, but also a corresponding influx of new community members who carry on the work. Departures frequently result in short-term challenges, but just as often, incoming community members present opportunities to reassess, reorient, and grow in new directions. Within scientific teams, there is sometimes tension between fulfilling our obligation to train early career scientists and patching holes that open in programs when talented technicians, graduate students and Ph.D.-level scientists actually leave to advance their careers elsewhere. But let’s go back to Blake Meyers.
Blake will be leaving soon to take the position of Director of the UC Davis Genome Center, which focuses faculty, core facilities, and “omics” approaches to address questions across the breadth of biology. While I’m excited to see Blake grow with this opportunity, I’m also pondering how much he and his team will be missed. Blake’s journey to the Danforth Center started at the 2014 American Society of Plant Biologists (ASPB) annual meeting in Portland, Oregon, when I asked him over dinner at Departure Restaurant, “What would it take to get you to the Danforth Center?” He arrived in 2016 with a sizable team as the first joint faculty hire with the University of Missouri – Columbia.
Since then, Blake and team have done pioneering research resulting in over 100 research papers, mostly focused on small RNAs in plants. They discovered the complexity, diversity, and functions of plant small RNA classes, such as those required for pollen formation in maize and other crops. The work revealed insights into evolution of plant genomes, mechanisms of disease resistance, and how plants reproduce, and has potential applications that include development of new hybrid crops. For these and numerous other contributions over the past 25 years, Blake was elected as a Member of the National Academy of Sciences in 2022.
We exist in world where people come and go. Nonetheless, the loss of Blake and team will be felt for quite some time. They are important contributors who have made the Danforth Center community better. But we’re a resilient Center, and bringing on board new faculty and team members in the months ahead are things we can anticipate with excitement.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Towards the end of 2024, we’ll celebrate the invaluable contributions of Hal Davies, VP for Finance and Chief Operating Officer, who has been bedrock at the Danforth Center since early 2001. Hal informed us that he will retire at year end, meaning he will depart as one of the two longest-serving members of the Danforth Center community.
When Hal first joined the Center as Controller, or lead accountant, he led a finance department with only one other person. Few systems and procedures were in place to manage grants, pay bills, plan budgets and handle the myriad complexities of finance. And there was the minor complication that Hal no grants experience! “I learned grants on my own,” Hal told me recently. “I was originally the grants compliance person, too.” All of the financial systems to run the Center in those early years were developed from the ground-up by Hal and his small team.
Working closely with Sam Fiorello, by 2011, Hal managed of a sustainable funding model for Center operations involving grants, annual donations and draws on our endowment. Along with ambitious fundraising plans, this allowed preparation for and execution of the Expansion Phase (2011-2020) in which we doubled the numbers of Center community members, grew the endowment and financial reserves, constructed the WHD Building, and built several new plant growth and technical facilities. Sustainable growth continues to this day.
I asked Hal what has been most surprising during his journey at the Danforth Center. He said, “Learning what plant scientist do, and how we make impact. Participating in the growth of the Center and the quality of science is personally fulfilling. Even though I’m not a scientist, I feel like I’ve contributed to the science.” Hal reflected further on what it’s meant to him personally to contribute to this organization. He said, “Being part of Bill Danforth’s vision and seeing that become a reality…it changed my life. I think I became a different person, realizing there is something bigger to achieve.”
I deeply appreciate Hal’s longstanding contributions to the Danforth Center, and his constant commitment to help our scientific and administrative teams succeed. I will communicate more about planning for a successor at a later date, but for now, let’s all thank Hal for a job well done!
Jim Carrington,
President and Chief Executive Officer
Bonus Content: On a personal level, I credit Hal for giving Teri and I one of the biggest thrills during our time in St. Louis. Hal used to own a set of four season tickets for great seats in section 347 at St. Louis Cardinals baseball games, and he would share the cost with friends who bought sets of tickets. For years, we bought two tickets for eight games. One time, we could not attend a game for which we held tickets, so Hal offered to trade for one of his game dates. What did we get in the exchange from Hal? Game 6 of the 2011 World Series, one of the greatest games in Cardinals and World Series history. Thanks, Hal!
Dear Danforth Center Community,
How about a roundup of recent happenings and events that are worth highlighting for the Danforth Center community? I’m going to start with something we’re taught never to lead with…
…an Apology. This past Monday, the St. Louis region experienced hazardous morning driving conditions due to icy roads. Just ask a local fire truck crew if you need a reminder! Due to concerns about safety, we closed the Danforth Center to non-essential personnel. But unlike schools and many other organizations on Monday, we failed to communicate the closure until 8:31 am. The late notice was distressing to many community members, including those who had already driven to the Center and those who had arranged child care prior to the announcement. I sincerely apologize for the late notification and aggravation this caused. We will review our inclement weather closure protocol, including how and when we notify the community, and make changes to ensure this does not happen again.
Core Facility Orientation. I heard and read great things about the Core Facility Orientation last week! This is a new, recurring program timed to coincide with new community member orientations. A primary purpose of the program is to inform existing and potential core facility users about the services provided and how to gain access. So, I was surprised to learn that the majority of attendees were from administrative and non-scientific departments. These attendees participated because they want to learn more about our scientific work, our technologies and our impact. This is such a positive reflection of how community members across the Center identify with the mission and want to learn about how our core facilities contribute to meaningful outcomes. Many thanks to each core facility team for delivering the event, and to all participants for attending.
Bioinformatics and Beer. Did you say you want to learn more? I hope to see you at Bioinformatics and Beer this afternoon (2:00 pm, AT&T Auditorium). Among the 15 regional speakers giving short talks on data science-intensive topics, five are from Danforth Center teams. The event is intended to build regional collegiality, collaboration and strength in data science-relevant research, and to get to know better our colleagues from Washington University, Saint Louis University, and other organizations over a refreshing beverage. I appreciate the good work from each of the speakers, organizers, event team members, and participants.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
High quality working space for Danforth Center teams is one of the most important, sought after, guarded, and sometimes envied features of our facilities. Unlike the cosmos, space at the Center has clear limits. While people, projects and priorities are dynamic, our physical working space is relatively slow to change. Let’s consider Danforth Center space, how space decisions are made, and what we do when space needs change.
How are space assignments made? For the Plant Growth Facility (PGF) or the Field Research Site (FRS), requests are submitted by PIs or teams and assigned based on availability. Space is assigned and managed by facility personnel through processes that may involve input from the facility oversight committee. Space allocations in PGF and FRS can grow or contract over time based on the needs; because retaining space has a cost, there is an economic incentive for teams to relinquish space that is no longer needed.
For laboratories and offices, spaces are assigned to research groups, departments and programs by the Space Committee, composed of me and Becky Bart (formerly Toni Kutchan) in our roles as President/CEO and VP for Research, respectively. Additional laboratory and sit-down space requests are common due to the addition of research team members brought on board with new funded projects. The Space Committee considers a number of factors when a request is made: What is the NEED, what is the AVAILABILITY of space, and what is REASONABLE in the broader context. The broader context includes competing needs, planning for future space assignments (e.g. for a new Assistant Member and team), and fairness. Sometimes the committee seeks to address the needs of multiple groups simultaneously. This is usually the case when an entire team is relocated to cover their needs, and the vacated spaces are used to address the needs of others. Recording and implementation of space assignments are managed well by Todd Hornburg as VP for Facilities.
What happens when critical needs are high but space is simply not available? With some careful planning, fundraising and budgeting, we can construct facilities like new greenhouses or the building extension that we’ve initiated adjacent to the PGF. We can also redesign existing space to better address current and foreseeable needs, like what was done to provide new instructional and training space.
Space can be a contentious issue, but I sincerely appreciate the professionalism and patience that I see regularly from Center community members who have needs.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Two of the most important types of investments we make at the Danforth Center involve recruiting new people and building new facilities. Let’s get everyone up-to-speed on recent developments in these two areas, each with important ramifications for our future.
Assistant Member/PI search. We recently activated a search for an Assistant Member/PI, who will build and lead a new research team. Bringing new research groups to the Center can have a big impact through introduction of new ideas, strengthening key priority areas, bridging important technical gaps, and broadening our network of collaborators. The search committee composed of Malia Gehan (Chair), Noah Fahlgren, Armando Bravo, Doug Allen, and Myia Elliott is just now beginning work.
Joint Mizzou-Danforth Center Faculty search. Our partnership with the University of Missouri – Columbia aims to have two joint faculty members based at Mizzou. A restart of the search for the second Mizzou-based joint faculty member is underway with a new committee, which includes Tessa Burch-Smith and Keith Slotkin. The search is focused on senior or mid-career candidates who qualify for appointments as both a tenured professor at the university and an Associate or Full Member at the Danforth Center. These cost-shared joint appointments aim to increase collaboration and partnerships, and to elevate plant science across the region.
New Building Construction. Speaking of hiring, the addition of new teams can put pressure on availability of space. We have experienced serious space limitations in recent years, including within the Plant Growth Facility (PGF). Despite the dramatic growth in size and scope of greenhouse and growth chamber facilities, and in the size of the PGF team needed to run the PGF, the amount of office, meeting and sit-down space for PGF team members is entirely insufficient; space for these purposes has remained static for over 20 years. We will solve that problem with construction of a 6500 sq. ft. building extension to the west side of the PGF headhouse area. In addition to the PGF team, the new space will house teams from the Phenotyping Core and Environmental and Health Safety/Biosafety. Construction begins next week.
I sincerely appreciate and thank everyone working on the new searches and the building project. Their efforts will have long-lasting impact on the Center! I wish everyone a thoughtful and meaningful Martin Luther King, Jr. Holiday on Monday.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Early January is an interesting time for some. Goals are set, self-reflection occurs, and New Year’s resolutions for self-improvement are made. I’ve never made a New Year’s resolution, though I wish all the best to those who do. This year, however, I did do something that I hope will benefit me and others in the Danforth Center community: I asked eight trusted Center colleagues, “What can I do better in 2024?” Here are the responses.
- Be better at communicating important things proactively or immediately
- Make Faculty Meetings more interesting
- Find better ways to make Faculty Meeting conversations more substantive
- Provide more clarity on how decisions are made
- Convene President’s Faculty Advisory Council (PFAC) more frequently
- Give more information about changes in the Leadership Team
- Just help me know what’s going on better
- Make sure everyone is recognized
- Help set up a comprehensive strategy for my team
- Join me for lunch some time!
While this is not a complete list of things you might think I can do better 2024, I do note a few themes. Most clearly, over half of the comments relate to doing a better job communicating about happenings, decisions, and achievements around the Danforth Center. There are a lot of sources of information that go to the Center community, and I commit to finding ways to better communicate news and other content that needs to, or should, come from me. Perhaps the Weekly Message can be used more effectively.
Two respondents zeroed right in on doing a better job using the time we have in Faculty Meetings. I can use some help in this regard, and I encourage attendees to provide ideas for content or discussions that are more relevant, engaging or necessary. One respondent surprised me with a suggestion that I could be more active in helping with strategic planning with their team. Although we have a Center-wide strategic plan through 2025, planning at the department or lab level may be just as important, or more so, for team members.
I sincerely thank all those who offered their input for how I can do better in 2024. And if anyone else has helpful suggestions, maybe we can have lunch together. I wish everyone a positive and productive 2024.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
While arriving and walking to my office the past two mornings, I’ve passed Chuck Maye and Chris Martin (Facilities team) working on installation of a new water bottle-filling station on the 3rd level. As I told them, I’ve had the same thought both days: “Wow, are you guys always here working hard?” Given that today is Friday, December 22 with the holidays at our doorstep, most of the Danforth Center community is taking advantage of well-earned vacation leave. Passing Ryan Delpercio with a cart full of soybean plants, Tira Jones carrying a full tray, and Ron Smithey moving quickly between work sites were reminders that Chuck and Chris are not the only ones so active on site today.
That leads me to conclude 2023 Weekly Messages with a few simple thoughts, starting with my sincere thank you to everyone who gives effort that is above and beyond, like those who serve essential functions or are on-call during weekends and holidays. Thank you to everyone who contributed so much to a productive year at the Danforth Center. We succeed because individuals and teams focus on their distinct roles and collaborate with others across the Center. And I thank everyone who takes the Danforth Center mission to heart and does their job with the belief and knowledge that real people in the world will benefit from their effort.
As I finish writing this message, I was just interrupted by Bill Stutz and Andrew Witthaus (Information Technology team) and Allison Brown asking how they could help with a meeting that I’ll be hosting in a few minutes. I am reminded again that it’s an honor to be surrounded by hard-working people who contribute their professional efforts so generously at the Danforth Center. Happy Holidays!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
There’s working quietly behind the scenes, and then there’s Ann Kruse! I suspect most of you have never met Ann. But since joining the Danforth Center in 2007, she has had an outsized but under-the-radar impact on our work, primarily as the manager of grants, budgets, consultancy agreements, and more for the multi-national Virus Resistant Cassava for Africa (VIRCA) project led by Nigel Taylor. The VIRCA project is delivering improved cassava varieties to small holder farmers in East Africa, first in Kenya, and has involved hundreds of scientists, trainees, communications specialists, regulatory experts and support teams from the U.S., Kenya, Uganda, Rwanda, Nigeria, and Canada.
Supporting an international project with multiple funding sources (currently Bill and Melinda Gates Foundation, U.S. Agency for International Development) is an enormous challenge requiring exceptional skills in organization, financial management and project management. Ann has been that skillful individual working stealthily, quickly, accurately and effectively to hold so much of the project together. She manages budgets for teams in multiple countries; she organizes international meetings and travel; she helps students and visiting scientists come to the Danforth Center. I cannot possibly list everything does, but I can say that she navigates all of her diverse functions with remarkable focus, patience, good humor. And when the VIRCA project manager (PM) left a few years ago, Ann stepped in and took over many of the PM roles on top of her numerous other responsibilities. “Without her I could not have kept the project running over this period,” Nigel recently told me.
Now, if you think my and Nigel’s opinions of Ann are high, you ought to hear what the international leads of the VIRCA partnership have to say about her! I recently read comments from seven such individuals. They praised Ann with terms including diligent, patient, meticulous, fantastic, genuinely caring, respectful, and “bedrock for VIRCA”. One of them said, “You have been a strong pillar for all, especially the partners and how you have managed us with so much professionalism and efficiency.” Ann has a lot of admirers overseas and at the Danforth Center because she’s helped so many succeed!
After 16 years and so much that she has achieved, Ann will retire from the Danforth Center next week. I am sad to see her leave, but so proud of what she’s done. I hope everyone joins me in expressing sincere appreciation for Ann and her commitment to the Danforth Center, the VIRCA project, and the numerous international partners with whom she has worked.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
A few weeks ago, our Board of Directors met for the third and final time in 2023. The work that the Board does affects the Center in countless ways, and the work by so many of you to communicate our purpose, plans and achievements to the Board makes a big difference. The great efforts to guide a tour or deliver a presentation to Directors, or to develop the Center budget for Board consideration, are just a few examples of contributions that are clearly noticed and appreciated by Directors. I thank everyone who helped our Directors do their work and better understand us in 2023.
Several important matters that affect the Danforth Center community were discussed and decided at the recent meeting. Here is a recap of a few highlights, some of which I’ll return to in the near future.
- Kirk Czymmek provided a wonderful overview of the Advanced Bioimaging Lab, including descriptions of new, bleeding-edge instrumentation and the breadth of research enabled for our scientists and partners.
- The 2024 Center operating budget of $46.3M, as well as the capital and non-operating budgets, were approved. The operating budget includes a 3.5% overall pool for merit increases.
- The 2023 Scientific Advisory Board (SAB) report was presented by Eric Ward, SAB Chair. The favorable report focused on the work presented by a subset of PIs and Core Facility Directors at the SAB annual meeting in September.
- I presented a report on 2023 usage and new construction at the Field Research Site. The research and achievements made by Center teams on 65 planted acres, and within working facilities under active development, were truly appreciated by the Board.
- And though you will hear more about this from me later, I want to let our community know the exciting news that Penny Pennington was elected as new Chair of the Board. Penny, who leads the brokerage and financial advisory firm, Edward Jones, will serve a five-year term as Chair. I am excited to work with her.
Finally, during the meeting and dinner afterwards, the Board expressed appreciation to the outgoing Chair, Todd Schnuck, for his impactful work since 2019. Todd has been instrumental in our success over the past five years, and he deserves our sincere thanks!
I will be traveling all of next week, so the next Weekly Message will be on
Dec. 15. Until then, take care.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I want to return to my October 11 message to the Danforth Center community in which I expressed deep concern for all those who were directly or indirectly impacted by the October 7 attacks in Israel and subsequent conflict. At the time, over 1600 people were either killed or kidnapped, including some with connections to Center community members. Since that message, an escalated war in Gaza and the West Bank has resulted in nearly 10-fold more deaths, mostly among Palestinians, and displacement of nearly 2 million people, again mostly Palestinians. This human tragedy is all the more staggering given the death and suffering of large numbers of innocent people, including young children. The geopolitical history underlying or influencing this tragic conflict offers little encouragement for the future.
As I’ve heard from many of you, the past six weeks have been difficult to process and comprehend. These events affect all of us, regardless of political, religious, or social views. I have spoken with community members who originate from Israel or Palestinian territories, or who identify with affected populations. These individuals carry a unique burden at this time, and are further pained by Islamophobia and anti-Semitism that has grown significantly in recent weeks. They and their families are changed by these events, just as they have been shaped by histories that extend long before the current conflict. And these community members are facing the uncertainty or fear about what comes next.
This message is written to recognize and support of all those within the Danforth Center community who are hurting, and to encourage the entire community to reach out to colleagues who might need a little help or just a sympathetic friend to talk with. These expressions of concern can make a big difference. Our values compel us to reach out and seek understanding about our diverse community members, especially in times of need. And for those in need of professional help, as most of us do from time to time, our Employee Assistance Program is there for you at no cost.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
We were pleased to hold the first Todd Mockler Memorial Lecture this week, a new annual event to remember Todd and to hear from a speaker who was uniquely impacted by him or his work. We were thrilled to honor Malia Gehan as the inaugural lecturer.
Malia spoke about her journey from being a postdoctoral fellow in Todd’s lab to a leader of her own team as a soon-to-be Associate Member and PI at the Danforth Center. She talked about how she learned from Todd to think bigger and attack important problems outside of her comfort zone. When the Bellwether Phenotyping system was installed 10 years ago, it was Malia and fellow postdoctorals Noah Fahlgren and Max Feldman who led development of new computer vision software, called PlantCV, to extract meaning from the voluminous data now on our hands. This was all the more impressive since Malia was not exactly what one would call a “skilled coder” or “accomplished programmer” at that time. But her determined drive to build computing tools that any plant scientist could use productively resulted in PlantCV becoming a widely adopted standard within the image-based phenotyping community. Malia’s highly collaborative research uses genomic, computational and other approaches to understand how plants deal with environmental stresses, like high temperature.
Malia had several concerns about this lecture ahead of time. First, she wanted to represent well those who have worked with Todd through the years; she was spectacular in doing just that. And second, she was uneasy about speaking in front of her everyday colleagues. “I was more nervous speaking in front of a Danforth Center audience than in front of an outside group,” she told me. I could relate to this. There’s something about speaking in front of a familiar crowd that raises anxiety, perhaps because you sense that they know too well exactly what you’re really like. That voice of the imposter inside our heads often gets louder as we approach a microphone in front of discerning associates.
I thank Malia for accepting the invitation to be the first Mockler Lecturer, and for representing so well her past influences, present achievements and future impact. She embodies the best of both the Mockler legacy and the Danforth Center community.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Let’s start with a big “Thank you” to everyone who participates in the broad range of donor-based or philanthropic fundraising that supports the Danforth Center and our strategic priorities. Though we have a talented and experienced team in Development that spearheads our fundraising endeavors, this is really a shared effort with every other team at the Center. Indeed, success in fundraising is shared success that benefits all corners of our organization.
Our fundraising priorities track closely to the priorities, initiatives and desired outcomes specified in the 2021-2025 Strategic Plan. Generally, fundraising from philanthropic sources enables us to construct and support facilities, develop core capabilities, undertake new initiatives, and do those things that cannot be easily supported through competitive grants from research funding agencies. Without philanthropic support, we would be unable to function as an institution. The non-philanthropic funding agencies (e.g. National Science Foundation), on the other hand, support research projects led by PIs and acquisition of some instrumentation.
Here are just a few of the many recent shared successes in raising support from donors.
- Support for Core Facilities – The Advanced Bioimaging Lab was able to acquire the Hydra Bio Plasma-FIB for ultra-high-resolution cellular cryo-tomography (for fun, repeat that quickly 5 times). We have initiated building of a new office, meeting and working space for PGF, Phenotyping, and Environmental Health and Safety teams. The Phenotyping Facility will soon start on both a major refurbishment of the Bellwether Phenotyping Facility and a significant new field phenotyping initiative.
- Development of Field Research Site – Speaking of the Field Site, most of the $11.2M for acquisition and development came from philanthropic donations. Construction of mechanical, storage and working facilities at the site is underway now.
- Support for Graduate Student Fellowships – We will be expanding support for graduate students with a new annual, named fellowship that will start in 2024.
- Authentic Research Experiences (pre-college) – This important program led by Education Research and Outreach has received philanthropic funding for student support, development of resources, and transportation.
These and all of the other recent fundraising successes have involved collaborative participation for critical conversations about needs, proposal planning and preparation, and cultivation and stewardship of donors. In some cases, you have stewarded and built trust with prospective supporters long before a gift is proposed. I and the Development team are deeply appreciative of all your shared efforts!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I met with the President’s Faculty Advisory Council (PFAC) yesterday to undertake the annual process of reviewing and revising faculty committee assignments. I know, that’s not the best hook to draw you in, but please keep reading. Though work on committees is necessary to cover important roles and responsibilities, like oversight of core facilities, for me it was an opportunity to reflect upon the following question: Are we expecting too much from our hard-working, over-achieving and sometimes over-stressed Danforth Center community members?
We have a large number of productive, ambitious, self-driven individuals who are achieving big things here. We have countless scientists, technical specialists, administrative personnel, facilities personnel, fundraisers, and other professionals who take on heavy lifts representing more than their fair share. Many have additional responsibilities or shared appointments with other institutions, or commitments beyond the Danforth Center. Some take on and achieve more as part of their intrinsic nature. Some are motivated to do more by our mission and vision, and the possibility of improving the lives of others. There are those who we’re over-assigning, or are self-overloading, with expectations that may exceed their capacity. And then there’s the important reality that some of our over-stretched community members may feel under-appreciated for the work they do.
Learning about how many of you feel about these issues in recent weeks and months has been helpful and, in some cases, eye-opening for me. It provided part of the motivation going into yesterday’s PFAC meeting to lessen the overall burden of committee assignments. Generally, we sought to shrink the size of committees that do not need large membership. We sought to limit the numbers of committees each person sits on, and to be more even-handed in balancing workloads. We also factored in the fact that many serve on committees or have important responsibilities at other institutions.
Of course, there is much more to consider beyond committee assignments. Are we over-assigning work to our most talented team members rather than reaching out to others who are capable but need a little help? Are we over-asking our best communicators to give tours and presentations rather than bringing along others who are willing but a little less experienced? I encourage all of our leaders and managers to give some additional thought to how we’re working with those who have more-than-full plates, what we’re asking of them, and how we can better express our appreciation.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
After the recent announcement about Toni Kutchan’s upcoming retirement at the end of this year, several of you have asked, “What will we do to fill the vacant Vice President for Research position?” I’ll get to the answer in just a moment, but first let’s consider what the Danforth Center VP for Research (VPR) actually does.
The VPR serves as the Center’s senior scientist and a key member of the Leadership Team. They are influential in setting strategic directions and priorities. The VPR’s roles and responsibilities partly include:
- Faculty Development – The VPR leads the Promotion and Retention process and mentoring program for faculty in the Member track. The VPR is instrumental in all new faculty hiring, and as a member of the Space Committee, helps decide how working spaces are allocated.
- Allocation of Special Funding – The VPR leads the processes to award competitive grants for new equipment, graduate student fellowships, and internal research projects.
- Research Compliance – The VPR oversees or participates in our programs for biosafety, environmental health and safety, research integrity, and management of regulated materials.
- Oversight of Core Facilities – Among other roles, the VPR oversees the directors for several core facilities.
In addition, when we experience surprises, emergencies or other unanticipated situations (e.g. a global pandemic), the VPR provides invaluable leadership that keeps the Center moving forward. While Toni deserves our thanks and appreciation for the great job she’s done in all of her roles, she deserves special recognition for her out-of-view contributions in dealing with various crises.
Moving forward upon Toni’s retirement, we will appoint an Interim VP for Research from within our senior faculty for 2024. The Interim appointment is intended to be of limited duration, though the expectations for this position holder to lead, contribute and help move us ahead remain high.
Oh, I almost forgot. Becky Bart has generously agreed to serve as the Interim VP for Research. Becky has been at the Danforth Center since 2013, leading an ambitious research program, directing the SINC Center, and serving in numerous other capacities. I’m thankful that Becky is willing to take on significant new responsibilities, and excited to work with her in this new role.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I hope everyone takes a few minutes to visit the recently installed feature that recognizes and honors those who lived on this site prior to the arrival of European and U.S. settlers. Overlooking the Danforth Center prairie near our main entrance, just east of the water garden, stands a plaque that reads:
As an institute focused on preserving and renewing the environment through plant science, the Donald Danforth Plant Science Center recognizes and honors Indigenous Peoples as the original stewards of the land. The Danforth Center acknowledges that it is located on the ancestral and unceded lands of the Osage, Missouri and Illini people, who were removed unjustly, and that we in this community are the beneficiaries of that removal. By recognizing Indigenous Peoples and their traditional homeland, we express gratitude for their enduring stewardship of the land. We honor and respect Indigenous Peoples, past and present, by building a more inclusive and equitable space for all.
Populations of indigenous nations and tribes that lived on this land were dynamic. The Osage, for example, originated from populations that migrated over a few centuries from the Ohio River valley towards the Mississippi River, eventually moving further west to become the dominant power in central and western Missouri by the early 19th century. The Illini were estimated to number approximately 10,000 people across 12 or 13 tribes (including the Cahokia in this region) along the Mississippi River valley in the 1600s. Due to conflict and disease, however, the Illini numbers were reduced to a few hundred by the late 1700s.
Starting in the early 1800s, the U.S. government instituted a series of policies that resulted in displacement of Native American populations. The Indian Removal Act of 1830 wrote into law that the government could legally take land from Native American nations and tribes, and force relocation to designated territories in the west, mainly in what is now Oklahoma. The Osage, Illini and other Indigenous Peoples from this region ultimately suffered this fate, as well as further harm when the U.S. failed to fulfill terms of agreements made during forced relocation.
Thank you to all of the Danforth Center community members and external partners who advocated for, facilitated, wrote, built, and installed this plaque that stands as a permanent reminder of the original inhabitants of our site.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
A few weeks ago, I wrote about the logic used for decision-making in my role at the Danforth Center. In retrospect, I undersold the fact the nearly all major decisions involve a collaborative effort of many people through information gathering, assessment, persuasion or consensus-building. Creation of the Danforth Center annual budget is a great example of such teamwork. The budget lays out anticipated revenue and expenses for the year, and once approved by the Board of Directors, serves as the financial blueprint for our work. We’re a not-for-profit organization; but don’t forget, we’re also a not-for-loss, so the budget needs to balance. We’re well underway in developing the 2024 budget, and I think it’s worth understanding what goes into that process.
Let’s start with anticipated revenue. The money that supports our salaries, supplies, and most everything else comes predominantly from two types of sources – competitive grants and philanthropy. Grants support most of the direct costs (e.g. lab supplies) and some indirect costs (e.g. facilities support) of research projects managed by PIs. Philanthropic gifts cover most other Danforth Center costs, and they enable us to function as an organization. As per the intention of each donor, some gifts are made to support annual mission-relevant activities, and some are given to grow our endowment. We draw an amount from the endowment each year by a formula. For the current 2023 budget, we planned for $19.1M in grant revenue, $2.4M in annual donor gifts, and $20.1M in draw from the endowment. We also planned for $5.9M from a variety of other sources (e.g. external user fees for core facilities), bringing the total anticipated funding level to $47.5M.
On the expense side of the budget, the leaders of each department submit a detailed budget request in the Fall for the upcoming year. Proposed spending is reviewed carefully with an eye toward meeting essential needs, supporting strategic priorities and good opportunities, financial sustainability and stewardship. Department leaders are appreciated collaborators in the budget process. For 2023, we budgeted for operating and non-operating costs totaling $46.3. The slight anticipated surplus at year-end will go into our operating reserve, which we maintain to cover unexpected events or future income shortfalls.
The budget planning process is managed, overseen, and executed with great skill by Djuan Coleman, Hal Davies, Leticia Slack, Darine Kube, Karla Elliot, and Heather Bowen in the Finance Team. Let’s all thank them for their hard work!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
It occurred to me that I’ve not highlighted any Danforth Center scientific publications recently. Let’s remedy that right here with three excellent papers, and with a few comments to help the non-specialists. Congratulations to all of the contributors!
Yu, Y., Hu, H., Voytas, D.F., Doust, A.N., and Kellogg, E.A. (2023). The YABBY gene
SHATTERING1 controls activation rather than patterning of the abscission zone in Setaria viridis. New Phytologist 240, 846-862. DOI: 10.1111/nph.19157.
Plants have highly evolved mechanisms to shed or drop organs (abscission) in response to environmental or developmental cues. It’s why you have to rake leaves in the Fall! But in agriculture, the natural shedding of flowers or seeds is detrimental, and cereal crops with abscission-inhibiting mutations in certain genes, like SHATTERING1, have been bred to dramatically increase yield. Using Crispr gene editing and detailed analyses, Yu et al. show for the first time some mechanistic features of abscission in the grasses, including the role of the plant hormone auxin.
Ludwig, E., Polydore, S., Berry, J., Sumner, J., Ficor, T., Agnew, E., Haine, K., Greenham, K., Fahlgren, N., Mocker, T.C., Gehan, M.A. (2023). Natural variation in Brachypodium distachyon responses to combined abiotic stresses. Plant J. DOI:10.1111/tpj.16387.
Understanding how native plants deal with extreme temperature, drought, and other environmental stresses offers insights into how crops can be bred to better withstand the harsh realities resulting from climate change. Ludwig and collaborators studied the genetic variation in how 149 different accessions of the grass Brachypodium distachyon from diverse Mediterranean and Middle East locations deal with heat, drought and combined heat+drought stresses. They found surprising relationships between heat and drought responses, and at least one new genetic determinant that controls the amount of stress damage.
Geng, S., Hamaji, T., Ferris, P.J., and Umen, J. A conserved RWP-RK transcription factor VSR1 controls gametic differentiation in volvocine algae. (2023). Proc. Natl. Acad. Sci. USA. DOI: 10.1073/pnas.2305099120.
This most recent work from the Umen lab gets at the critical control mechanism for sex determination in single-celled and multicellular algae. The capacity to produce distinct mating types (e.g. male and female) is the foundation for reshuffling of genetic material within a species, which maintains genetic diversity and capacity to adapt in different environments. Geng et al. discovered that the key regulatory protein VSR1 either interacts with itself (VSR1-VSR1 complex) to induce female gametes, or with MID protein (VSR1-MID) to induce male gametes. The data show elegantly why either male or female gametes form, and they give reasons why this mechanism may also occur throughout the plant kingdom.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
As I’ve indicated often in the past, I spend a lot of time speaking about the Danforth Center and our work with public, non-scientific audiences. Largely through trial and failure, minimal formal training, a lot of observation, and more analysis than you would ever guess, I enjoy speaking to and interacting with public groups. And I have fun doing so, especially bringing the human element into the conversation about what we do.
Which brings me to the first 10 minutes of a Story Collider podcast episode called “Food Science: Stories About Things We Eat,” featuring Katie Murphy, Director of the Center’s Phenotyping Core Facility and PI. Katie talks about her unpredictable journey becoming a TikTok star, bringing the joy, excitement, inclusiveness and magic of plant science to the fickle, impatient social media audience. Like almost nothing else I’ve heard, Katie’s story shows what happens when scientists let the public in on the mysteries to be solved and surprises to be learned when we use science to answer a question. With humor, kindness, wonder and accessibility, Katie shows through TikTok and the podcast what most non-scientists rarely hear or see: life in the lab is funny, annoying, interesting, boring, surprising and quirky. That is, Katie shows us that a life in science is a very human, relatable life, and anything but a separate existence of elites looking to impose their will on society.
I think spreading this idea about the human nature of science, as Katie shows, has enormous power to break down barriers that have grown between scientists and much of the public. But we have too few people within the scientific community who are willing, capable and brave enough to face the public, armed with the amusing imperfections of a scientist, and risk criticism, hostility or indifference. I believe it’s exactly those amusing imperfections that scientists need to lean on to reach and relate to broader audiences. I think that’s a big part of what Katie’s outreach is actually doing.
I hereby deem Katie’s podcast required listening for everyone who talks to others about science or the work we do. And do your own followers a favor by passing it along. Have a good weekend.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
At a monthly meeting today for a collaborative project between the Carrington and Bart labs, much time was spent discussing project priorities. There are not enough people, resources, funding, space or time to do everything optimally for the project. Prioritization is a necessity. But because the nature of future results is rarely clear, prioritization intrinsically involves the risk of making ‘wrong’ choices, even though the choices today might seem sensible.
Prioritization and decision-making with uncertain consequences are major sources of worry in my role at the Danforth Center. Most prioritization decisions involve allocation or reallocation of funding, space, or peoples’ time and effort. With a few questions and real examples, let me share the process I use in making decisions.
- Is this a ‘must do?’ Decisions to get the ‘must dos’ done are often the easiest. We must maintain our facilities in working order, or comply with federal requirements. We must provide ASL interpreters to support our Deaf community members. Funding will always be allocated as a budget priority, or identified in the case of unbudgeted emergencies.
- Is this a strategic priority for which we’ve planned? Priorities in the 2021-2025 Strategic Plan are baked into our budgeting and fundraising plans. Some of these priorities, like development of the Field Research Site, are expensive, multi-year commitments, and we match up the pace of spending with the pace of fundraising. The open decisions are often about when to start.
- If this is a good idea outside of the strategic priorities but worthwhile doing, can we afford it? Here’s where we sometimes have interesting or difficult conversations, and often the answer is ‘no,’ or ‘not this year.’ But really good ideas are worth talking about because we may be able to apply for new grants from funding agencies, or secure other sources of novel funding.
- For each of the categories above, what are the collateral consequences? If we prioritize a Field Research Site, how does that affect future Center budgets? In support of Deaf community members, to whom are new responsibilities assigned and how do we rebalance workloads? If we assign additional lab space to a team, how will that affect our ability to recruit new teams? Broadly, how does a decision in one area affect others? What might seem like unnecessary delays in decision making is often because of the need to anticipate and plan for the collateral consequences.
My goal in all decision-making is not perfection, but to be reasonable. I thank all those who collaborate with me to achieve that goal.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
“How did they do that?” This was a question I heard immediately after the Big Ideas 3.0 presentations last evening. The program featured three teams of three early-career, Danforth Center scientists or educators. In front of a public audience of over 350, and 300 more online, each team presented a 14-minute talk about an ambitious, impactful idea that extends from their work, and then fielded questions from three panelists (thanks to Janet Wilding, Benjamin Akande and Marilyn Bush, and to MC Chip Lerwick) who had no prior exposure to the topics at hand. Each team performed brilliantly with engaging, audience-inspiring flair. But, how did they do that?
Part of the answer is rather obvious. Over 11 weeks, they worked incredibly hard on collaboratively formulating their ideas, and then organizing, agonizing, re-organizing, refining and rehearsing their presentations. At weekly convenings with a coach, each team also helped the other teams improve their talks. No one witnessing this would have guessed it was a competition!
But the other part of the answer is less obvious: they learned and practiced the big secrets about connecting with an audience. In addition to their accessible, plant science-based content, they incorporated anticipation, surprise, emotion, humor, movement, interaction, and other verbal and non-verbal tools to reach every corner of the audience. Early in the 11 weeks, they learned by studying and dissecting what all of the great performers, including singers, bands and stand-up comics, do onstage to engage an audience. It turns out that Louis Connelly saying, “I still find myself asking, what are plants made of?” can be just as compelling as Bruce Springsteen belting out, “Roll down your window and let the wind blow back your hair” if they’re using the same tools to connect with an audience. With practice and preparation, the result in both cases is the magic of sprezzatura and a special bond with the audience.
I will forever admire, and be grateful for, the nine talented Big Ideas 3.0 speakers: Poonam Jyoti, Somnath Koley and Stewart Morley (Team EcoTAG from Doug Allen lab); Antonio Brazelton, Zach Stafford and Kurly Taylor (Team Whatley from JJK-FAN and Kris Callis-Duehl team); and Louis Connelly, Britney Millman and Allen Hubbard (Team Metablify from Ivan Baxter lab). They were amazing! Along with behind-the-scenes sprezzatura from the Development, Events, IT, Facilities and PR teams, and Allison Brown, I cannot imagine a better representation of the Danforth Center and our community!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
It’s a good idea to occasionally ask yourself, “Who am I, and what am I doing?” I suppose you could interpret and answer these questions in a number of ways depending on your motivations, like a desire to justify one’s actions or alter a career path. I’m frequently asked variations of, “What is the Danforth Center, and why are we doing what we do?” These questions come from first-time visitors to long-serving Center community members. Let’s consider two variations of these questions that I’ve been asked recently.
“We’re a plant science discovery organization, so why worry about all this other stuff?” By “other stuff,” the questioner meant things like spinning out companies and outreach to the community. We were founded on a vision of plant science discovery leading to positive impact for food, the environment, and the St. Louis region. The Danforth Center invests in people and facilities to discover important things about how plants function and interact with the environment, but we also invest in moving discoveries out of the Center and into the hands of organizations and people who can benefit. Those benefits come in many forms, from products with better sustainability properties made by companies in 39North, to a better trained or enabled workforce. We are, indeed, a discovery organization, but we’re also a scientific engine that intentionally connects to other parts within our region and beyond.
“I thought the Center’s purpose was to help smallholder farmers around the world, so why not focus only on that?” In fact, our founders had a close eye on the benefits that smallholder farmers and their communities could gain by more access to scientific advances, and we’re committed to doing so through translational and product-focused work of IICI and lab groups around the Center. But we were created with the broader idea that scientific discovery from both applied and basic researchers would unlock possibilities for smallholder farmers and for other domestic and international beneficiaries. Bill Danforth was inspired by how scientists studying esoteric-sounding properties of viruses, like buoyant density and Svedberg values, enabled breakthroughs that led to the poliovirus vaccines in the 1950s. Center discoveries in a wide range of areas, from epigenetics to gene editing, are continuously used to explore and identify new ways to apply our science.
We are a combination of talents, interests and motivations, and we work on many different things. But unified around mission and vision, who we are and what we do are big parts of what makes the Danforth Center unique.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Contrary to common wisdom, the “vacation month” of August always seems as busy as ever around the Danforth Center. On top of the everyday non-stop activity, many community members are cramming to get work done ahead of upcoming time off. That’s me today. But before I head out tomorrow for a week of cooler clime and remarkable scenery, I want to recognize some of the good work I’ve witnessed in recent days.
InfoEd implementation. This complex, difficult initiative to deploy a new grants management system is nearing completion. Kinks are being ironed out, existing grant data are being uploaded, and training of users is underway. I thank Melissa Kerckhoff for leading the implementation effort, Michelle Richards for excellent training, and everyone who has helped integrate InfoEd into our financial, proposal submission, sponsored projects, and other systems.
Great events. It was nice to participate in this week’s inaugural Ignite event, which seeks to bring people together from the region and inspire actions that align with our mission. Thanks to the numerous individuals and departments, led by Stephanie Regagnon and the Innovation team, that collaborated to raise funds for, organize and deliver Ignite. Next up: Big Ideas 3.0 on August 31.
Plant growth facility and field site management. Plants in the Plant Growth Facilities and Field Site do not get an August break. That means we owe a deep debt of gratitude to the managers, horticulturalists, farm team members and others who care for the invaluable plants in our experiments through the hottest time of the year.
Donuts and Donations. Many thanks to the Development team for hosting a donut-packed tea time to celebrate the 130 donors within the Danforth Center community this week. Pat Baldrich and Katie Murphy explained why they contribute financially to the Center, how those donations are used, and the importance of high donor support from within. Thank you Pat, Katie, and all of our in-house donors!
I’ll write again in two weeks after returning from where the water tastes like wine and the climate suits my clothes. Take care.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Ten weeks of hard work. Twenty-one undergraduate student researchers. Twenty-seven mentors. Incalculable career-long benefits.
For the 23rd time, the Danforth Center has hosted a talented, motivated cohort of students in the Summer Research Experiences for Undergraduates (REU) program. Though they are science majors at colleges and universities around the country, most REU students arrive with very limited or no experience in a research environment. Nevertheless, each student is immersed quickly in the deep end of the research pool. The fact that they achieve so much, as shown today during their end-of-program symposium presentations, reveals both their newly developed talents and their intense work. But that success also depends on some of the hidden REU program heroes – the mentors.
An REU mentor is usually a Danforth Center graduate student, postdoctoral scientist or senior research scientist who guides a student through the program. They help formulate a project, train in research methods, help collect and interpret data, and advise on all aspect of the REU experience. They do this on top of their full-time responsibilities, meaning they are doing something extraordinary. Dan Lin has been an REU mentor for six different students since 2016. He is a model of commitment, consistency, and compassion in this role, and the students with whom he has worked have achieved great things. I asked Dan what makes a good REU student mentor, and what he’s learned over the years? He had three things to say.
- “Mentors need to understand that students come from diverse backgrounds and experiences and with varied personalities and needs. Expectations need to be explicit, reasonable and aligned.”
- “Mentors need to be engaged and invested in the success of their mentees and adaptable in the face of challenges.”
- “Trust is crucial for a good mentor-mentee relationship; a good mentor needs to know how to foster and maintain this.”
It’s no wonder that Dan succeeds as a mentor year after year, as do many others who take on this important responsibility. In addition to REU program directors and coordinators Tessa Burch-Smith, Kirk Czymmek, Judy Mitchell and Monica Alsup, I thank all of the mentors for investing time and energy to “pass the baton” to the next generation. In the years ahead when our current REU students are mentors themselves, the lessons they are living through with their Danforth Center mentors today will be remembered and passed along again.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Several of us boarded a bus with eight Board Directors to take a trip out to the Danforth Center Field Research Site this week. The Board approved acquisition of the 140-acre property last year, and has since seen numerous presentations about site development plans and progress. What most of our Directors had yet to see was the actual work underway at the new facility. Our goal was to immerse the Directors and accompanying Center community members in the purpose, ongoing research and teams at the Field Site. The secondary objective was to survive the heat anticipated on the hottest day of the year.
In front of multi-year trial plots of intermediate wheatgrass, or Kernza, they heard from Molly Hanlon and Matthew Rubin about research to improve yields and sustainability benefits of perennial grain crops. Daniel Mullins showed how he and Molly are continuously imaging and measuring perennial plant roots using mini-rhizotrons. At a plot with diverse sunflower varieties, Toni Kutchan talked about research to identify new, high-yielding sources of natural rubber. Allison Miller and colleagues from Saint Louis University presented our Taylor Geospatial Institute collaborations around drone-based phenotyping to monitor plant performance across the entire Field Site.
Under a welcoming shade tent, Getu Duguma showed the first field trial of short-stature tef developed at the Danforth Center. Shorter varieties are needed to prevent lodging (plants falling over from wind and rain) in geographies like Ethiopia where tef is a staple; everyone could see dramatic evidence of the benefit of Getu’s work. Andrea Eveland then walked us through the Maize Maze, an education and outreach resource to demonstrate the wonderful world of corn genetics. After comments in Mockler House (Field Site headquarters) on site development, perennial roots and cover crop research from Katie Murphy, Allison Miller and Chris Topp, and the fabulous gift of Kernza brownies from Matthew Rubin, we were back on the bus for the ride back to the Center. The entire trip was a masterclass in organization and coordination.
As several Directors mentioned during the Board meeting later that afternoon, it was so exciting to see the Center’s work extending out of the lab by talented people with so much enthusiasm and purpose. I thank everyone who made this visit a reality, including all those mentioned above, Diane Moleski, and the entire Field Research Site team of Terry Beeler, Nelson Curran, Kevin Hava and Jenna Wood. You each demonstrated the very best of who we are!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
For years, Allison Brown (Executive Assistant in my office) has been asking, “When can I get rid of the 10 or so boxes of reprints of your old papers?” Each box weighs in at around 30 pounds and sits in a secret closet up on the 3rd level.
I mentioned this to a group in my lab recently and got the following question: “What is a reprint?”
For the younger scientists, reprints were physical paper copies of individual research articles that you received from the journal after a paper was published in the old days. By that I mean before scientific articles were widely available through open-access or subscription online. Here’s how it worked in, say, the 1980s. When you published a paper, the journal would send a bill for publication fees, typically a few thousand dollars, with extra charges for each fancy color image. Long papers costed more. They would also send a “Reprint Request Form” for you to request official reprints at a price of $10 or more each, provided that you bought a minimum of 50-100. By now, the younger scientists should be asking, “What the…?” and “Why in the world…?”
Here's how it worked. Scientists used to go to libraries, buildings that had books and scientific journals sitting on shelves. They would open the journals, one by one, and look for articles of interest. A not-insignificant number of scientists would also bring a stack of “Reprint Request Cards,” which were little postcards that you would mail to corresponding authors to ask that they send an official reprint of a specific paper. Upon receipt of the card, the author would mail a reprint to the requestor, whereupon the requester would possibly read it but definitely file it in a cabinet. Many authors would count, and save, the reprint requests cards they received, using them to gauge how much their colleagues were interested in the paper. Yes, before download metrics, likes, hearts and thumbs-up, we had reprint request cards!
Now you may be asking, since copy machines have been around since 1949, why didn’t people just photocopy the desired articles? The answer to that involves a combination copyright law, publisher greed, and mystery. All I know is now I have a few hundred pounds of non-requested reprints that I’ve been lugging around since 2005. But if you want one, you now know what to do.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Many thanks to everyone who came out last Wednesday to celebrate the Danforth Center being named a “2023 Top Workplace” in St. Louis. The basis for selection for a third straight year was our annual engagement survey results, which we use to assess how the community feels about the Center, our working environment, and our culture. According to survey data, the top-scoring culture drivers at the Center this year are:
- Development (Managers help community members learn and grow)
- Interdepartmental Cooperation (Different teams work together well)
- Open Mindedness (Different points of view are encouraged)
It is gratifying to see these three aspects of our culture ranked highest by the community. It means we come together, recognize our unique talents and perspectives, and help one another. This was on full display this morning during a lengthy collaborative team meeting between members of Bart and Carrington labs working on a Gates Foundation-funded project (Epigenetics in Cassava). I was deeply impressed by how Becky Bart (Lead PI) guided the entire team to diagnose challenges, think creatively, and design solutions that draw upon the unique talents and perspectives within the team. The meeting was all about how we could better help one another be more effective contributors to the project. In the process, I saw several team members, like Myia Elliott and James Rauschendorfer, embrace the opportunity to grow in their roles.
Then there are instances of community members helping each other spontaneously every day in ways that might seem small. Yesterday, I was with a small group in the Riney Family Greenhouse complex (B-range), looking to record some video for the upcoming Big Ideas event. My lack-of-planning failure to arrange for someone to give us access to a specific room meant that we encountered only locked rooms as we wandered the corridor. Then I met Eric Petit (Horticulturalist in the Plant Growth Facility), who approached and asked most politely, “Can I help you?” I explained what we were hoping to do, and within a few seconds he guided us to the one unlocked room and helped arrange some space for the brief video shoot. Thank you, Eric!
Helping one another in “small” ways makes a big difference. Lending a hand builds trust, confidence and appreciation between team members, cooperators, and those who are simply crossing paths. It’s a big reason we are a Top Workplace!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
“If I kept my front yard like you keep yours, I’d get fined by the city!” Starting in 2016, the year we seeded the Danforth Center prairie with 70+ native plant species, we received numerous phone calls like this from angry and appalled local residents, puzzled by what looked like a disorganized mess on our property. And we still get the occasional call. Today, with the prairie displaying the latter stages of a spectacular, month-long bloom of Rudbeckia and Coreopsis species, it’s hard to imagine anyone complaining. Because so many Danforth Center community members were not here when we transformed our landscape, let’s talk about the prairie and why it was developed.
Prior to construction of the William H. Danforth wing, the front landscape was a typical corporate grass lawn with ordered shrubbery and rows of ornamental pear trees. While the landscaping drew little attention, it was hardly a reflection of a creative plant science institution. Reconstruction of a Missouri tall-grass prairie, which dominated this region for 10,000 years after the glaciers receded, was based on several ideas:
- To honor the vast importance of native landscapes and natural diversity in the world;
- To acknowledge that native plants are the ultimate genetic base for all cultivated crops;
- To show that nature has already invented ways for plants to deal with diseases, pests, temperature extremes, droughts and floods;
- To maintain a far more environmentally sustainable property that sequesters more carbon, retains rainwater on site, recycles nutrients, and provides habitat for wildlife.
At the same time that the prairie was seeded, the pond at the front entrance was converted from a blue pool with a fountain to a living water feature containing aquatic plants. The bridge spanning the water was intended to be a functional walkway leading to the prairie and a symbol of the connection between the natural world and the research we do in our facilities.
The prairie is also one of our most important tools to inform and inspire the public about the work we do. From the tour route with interpretive signage to helping audiences understand how perennials work, the prairie has many “touchpoints” that we can all use when interacting with the broader community. I now get much enjoyment when I hear a local resident tell me, “I had no idea what you were doing on your site, but I get it now. It’s amazing!”
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Several community members have asked if the Danforth Center will be affected by the recent Supreme Court decision that will nullify Affirmative Action programs in higher education. As I usually try to do in these messages, I’ll seek to avoid politics or out-of-my-lane commentary on Constitutional law. Let’s consider both direct effects and indirect effects of the Court’s decision on the Danforth Center.
The Danforth Center will not be affected directly by this decision. The ruling applies only to Affirmative Action programs at colleges and universities. These programs include those intended to promote a more diverse student body. According to Charlotte A. Burrows, Chair of the U.S. Equal Opportunity Employment Commission, the ruling “does not address employer efforts to foster diverse and inclusive workforces or to engage the talents of all qualified workers, regardless of their background. It remains lawful for employers to implement diversity, equity, inclusion, and accessibility programs that seek to ensure workers of all backgrounds are afforded equal opportunity in the workplace.”
But because we partner with colleges and universities for training students, and we hire graduates of colleges and universities on a regular basis, the Danforth Center may be indirectly affected by the ruling. If the diversity of students admitted to our partner universities goes down, so will our ability to attract diverse undergraduate and graduate students to the Center. If future graduating classes are less diverse, the pools from which we seek to hire new community members will be less diverse as well. Over time, these may negatively impact our strategic goal to attract more individuals from historically underrepresented groups to the Center and STEM careers.
I am concerned about the long-term, indirect impact this ruling may have on us. Data from real studies show that a diverse workplace is more productive, has more satisfied employees, is more innovative and creative, and has a more positive culture. Irrespective of our individual opinions on Affirmative Action programs or Supreme Court decisions, I hope we can all agree that a diverse and included Danforth Center community is worth our continued, collective efforts.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
A majority of community members recently participated in our annual engagement survey, one of several tools we use to assess how the community is feeling about life at the Center. The 460 written comments were particularly informative; I read each one. Most comments revealed positive feelings, but let’s look at a sample of both favorable (+) and less-than-favorable (-) comments across five of the survey themes.
Values and Culture
(+) “How the organization lives the mission and values…many organizations have great missions and visions, but they are not integrated into the culture. However, at Danforth Center these are integrated into the day-to-day work.”
(-) “When I make suggestions for cultural change and/or general operation improvements, I have felt resistance and hesitancy to that suggestion. Feeling like my experience is not valued/appreciated because I don't have a higher degree…”
Collaboration
(+) “I'm genuinely impressed by the cooperation between all departments.”
(-) “Research/lab departments could do better with respectfully communicating with PGF and other departments. Listening to others about proper practices and use of space, following rules, and using common sense goes a long way.”
Efficiency and Communication
(+) “Danforth center does meetings, gatherings, and communications efficiently and well.”
(-) “Teams don't pass information along well. People in some areas aren't included in planning that involves their jobs.”
Senior Managers
(+) “Strong emphasis on regular communication through multiple platforms. Lots of outreach to community and stakeholders, not only community members.”
(-) “Lack of explanations make us not trust them. And, I think the senior management could maybe do a better job informing individual labs about the general direction of the center.”
Overall Comments
(+) “I work with great people who try their best. The resources available are the best available anywhere in the world. The science is creative, cutting-edge and expanding. I get a chance to try new ideas and take risks.”
(-) “When asked what ideas we have, or we are forced to do surveys like this…We can bring ideas to the table all day, but most are never accepted or just fall on deaf ears because what we say is not what some people want to hear. What is the point of this survey?”
I very much appreciate the entire range of feedback. While our community overall feels pretty good, not everyone feels included, heard or appreciated. Learning where we have concerns, and then working with leaders and managers to address issues within their teams, is precisely the point of the survey.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
We are concluding an interesting week at the Danforth Center featuring numerous activities and events under the theme, Threads of Culture: The Power of Story. Community members were welcomed at gatherings - sometimes large and sometimes small – to share our family and cultural traditions, our different foods, our unique celebrations, and the stories behind who we are. I participated in the community potluck lunch, a discussion about our names (and families), and several cultural performances. As Jennifer McDonnell in People and Culture recently told me, “The purpose behind this week was meant to share the commonalities that we have within our individual cultures and life experiences…these are aspects of our community members that we may not know about, but they are a part of who they are and how they show up.” With so many cultures represented at the Center, I found this to be a brilliant way to celebrate and encourage understanding about all of us.
The notion that we have commonalities across different cultures and traditions is really worth thinking about. We have so much intersection of values, needs, ambitions and purpose regardless of whether one comes from Redondo Beach, CA or Rubongi, Uganda. We stand united around the Center’s mission, for example, even though we originate from nearly 30 different countries. We work hard and do our jobs with diligence regardless of our sexual orientation or what we prefer to be called. We seek the best for our families regardless of our racial heritage. I appreciate how elements of unity came through this week.
Too often, our perception of individual differences interfere with understanding about what or how our other community members are actually contributing. What might be perceived by a less-than-fully-aware observer as meek or non-assertive behavior of a colleague may, in fact, be careful strategic thought and productive patience. I’m guilty of making erroneous judgments about people based on such superficial observations.
Weeks like this one at the Danforth Center should encourage us to continue seeking ways to find unity through our differences. Special thanks go to the entire People and Culture team and the DEI Council for their ideas and efforts to bring Threads of Culture to life. I wish everyone an enjoyable and thoughtful Juneteenth holiday on Monday!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
This message is being written while listening to fantastic presentations from Danforth Center scientists at the annual Scientific Retreat. I’m struck by how much this event has changed since the first one I attended in 2011. The Scientific Retreat today is attended by over twice as many community members compared to 12 years ago. The quality and scope of oral and poster presentations have grown dramatically over the years. The style of oral presentations have changed, now emphasizing shorter, snappier talks that are more accessible to the broad audience. We now have ASL interpreters for our deaf community members. And the location is different. Rather than an offsite retreat at Pere Marquette or Trout Lodge, this year we are using facilities at the Missouri Botanical Gardens and Danforth Center on consecutive days.
The post-pandemic decision to stay close to home rather than travel to an overnight location was not due to the sometimes-too-rustic facilities, the cafeteria cuisine, or the hungry ticks in Potosi. Instead, staying in town was intended to make the Retreat more inclusive and accessible to as many as possible. This includes parents and caregivers with limited travel capacity, and those who need to be onsite for their jobs. The recently constructed conference facilities at the Botanical Gardens offer an interesting new venue for a more inclusive offsite portion of the Retreat.
We’ve put a lot of thought into making the annual Scientific Retreat more respectful, safe and welcoming to our diverse community. How can we make it better? Pre-pandemic, the overnight retreat featured evening social activities, like karaoke. Turns out that for some, the life they love is making music with their friends. What can we do in the future that is fun, inclusive and promoting of a more cohesive community? The retreat organizers (Armando Bravo, Meter Nusinow, Kerri Gilbert) will welcome feedback.
Finally, many thanks to all those who make the Scientific Retreat successful, including the speakers, poster presenters, organizers, attendees, and administrative, AV, catering and facilities teams. Now, I need to get back to the talks.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
A frequent conversation topic with people who are learning about the Danforth Center concerns how we realize impact from our research. I sometimes talk about the rationale for fundamental research, the intrinsic value of scientific discovery, and our critical role in training scientists and STEM professions. But often, they are more interested in how our research and discoveries lead to new products and services that address important problems. And those conversations frequently include everyone’s favorite topic – Intellectual Property (IP), most commonly patents.
Why are patents necessary for the Danforth Center? If we make discoveries or develop technologies (inventions) with potential to contribute to more sustainable agriculture, more productive crops, higher value crops, or other benefits, we will file patents to “protect” those inventions. A patent give us the right to exclude others from making, using, or selling the invention for a limited period of time. While to some that might sound counter to our non-profit mission to improve the human condition through plant science, patents are a critical part of the route that inventions must take before they are used in the marketplace. The Danforth Center licenses our patents to companies big and small, including start-ups that spin out of the Center. Without those patents and the assurance of the right to commercialize, companies will not touch our inventions. And if our inventions do not reach the marketplace, farmers or other consumers will not benefit.
What about humanitarian uses of our patented discoveries and technologies? For these uses, the Danforth Center will grant royalty-free licenses. This is important when we engage in product development to benefit smallholder farmers in developing countries.
How do we go about filing patents? We are fortunate to have Erica Agnew (Director, Intellectual Property and Technology Management), a talented scientist and attorney, within the Danforth Center community. She has a law degree from Florida State University and, since 2021, has been a registered Patent Attorney with the United States Patent and Trademark Office. Erica works with Center scientists to help identify patentable inventions, facilitate and coordinate patent applications, and manage our portfolio of IP. She also participates in negotiations with companies, helps craft licensing agreements, and works with the leadership team on overall IP strategy.
Erica adds great value to the work we do, and is an essential contributor towards making impact at the Danforth Center. Thank you, Erica!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
If you participated in PlantTech Jam last Saturday, you were part of a significant event at the Danforth Center. We welcomed 563 guests from across the region with a wide range of hands-on experiences, geared primarily for kids. As a more plant-centered event that evolved from the former Raspberry Pi Jam, PlantTech Jam was a big success!
I was deeply impressed by the broad participation from Danforth Center teams that organized interactive tables, the inventiveness of participating partner organizations, and the large numbers of Center and external volunteers that helped with tours, soldering and much more. The creativity on display at the team tables was remarkable. Every station provided active engagement with Danforth Center scientists around a scientific principle or idea. Data show that participation-type experiences, like those offered at PlantTech Jam, improve students’ science process skills, STEM career interests, and motivation for STEM fields compared to the students lacking such experiences. Kids start identifying as scientists when that connect with real scientists around meaningful, inquiry-based activities. PlantTech Jam is something we can do uniquely to inspire future scientists and STEM professionals in St. Louis. Special thanks go to the organizers (Kris Callis-Duehl, Malia Gehan, Meter Nusinow, Noah Fahlgren, Jenny Nguyen, Cat Currens and Tam McGuire) and the many dozens of Danforth Center community members who were involved!
Other interesting opportunities for public engagement are on the horizon. One that I’m particularly excited about is Big Ideas 3.0 on August 31. Three teams of three scientists from the Center will compete by presenting a solution to a big challenge, based on their current work, in front of a public audience. If you’ve not seen either of the two prior Big Ideas events, think in terms of TED Talk meets Shark Tank. This is an exceptional platform to showcase nine young, talented Center scientists. And as shown previously, the big ideas can stimulate new thinking that turns into major focus areas within the team labs.
I’m also looking forward to seeing public outreach at the new Field Research Site, where the Eveland lab is creating a “maize genome tour” using a chromosomally ordered series of dramatic, colorful maize mutants. We all have unique talents and ways in which we can reach out beyond the Center’s doors. Let’s keep using them!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Two things of note happened around the same time 33 years ago. First, Gus Vogt, a Senior Laboratory Technician in the Meyers lab, was born. And second, now 86-year old Bobbie Miller moved into her second-floor apartment unit in which she has lived ever since. The two met for the first time last Friday evening, shortly after Gus heard screaming from the direction of Bobbie’s apartment building next to where he lives.
Gus quickly realized that the neighboring building was on fire. Panicked residents were running out to the front, and Gus immediately went over to help. When he arrived, Gus could see that the fire was located in the middle of the second floor. He quickly asked, “Are there people inside?” The short response was, “Yes.”
With few thoughts other than a compulsion to help, Gus entered the burning building. There were staircases at both ends of the 12 or so units on each floor. He ascended to the second level at one end and encountered thick, pitch-black smoke that completely obscured visibility above six inches from the floor. He crawled down the hall, banging on doors and calling out to anyone who might respond. He felt the heat of the fire, which prevented Gus from traversing the entire hallway. Without finding anyone, Gus crawled back and went downstairs. He then went up the stairs on the other side!
As he crawled to the first unit on the right, Gus saw the door was opened slightly. In the living room inside, he encountered a scared but conscious Bobbie Miller lying face-down on the ground, holding a wet towel over her mouth. Gus quickly helped her up, and with a combination of urgent assistance and forceful dragging, got Bobbie down the stairs and out of the building safely. When they exited, the stunned man who lived across the hall from Bobbie asked Gus, “Are you an angel?” Others asked, “Who are you…where did you come from?” Soon after, the firefighters and emergency responders arrived. And soon after that, with no one other than Bobbie and those who witnessed his heroism knowing what he had done, Gus was back in his apartment and ready to call it a night.
Each week, I look forward to writing about the wonderful achievements of Danforth Center community members working to deliver on our mission. This week, I am honored to recognize and thank Gus for his remarkable display of humanity, shown at great risk to himself and in service to people he had never met.
Jim Carrington,
President and Chief Executive Officer
P.S. Bobbie suffered little if any injury and is doing well. Bobbie and her fellow apartment residents were provided temporary housing and assistance from the Red Cross. As of today, no media or public safety officials have contacted Gus.
Dear Danforth Center Community,
When you organize an event, there’s nothing better than a packed house! Whether it’s to teach, inspire or celebrate, a full room of willing attendees adds incalculable energy and enthusiasm. I want to recognize four events that were at or near “standing room only” in the Langenberg Theater over the past week.
Kiona Elliott’s Thesis Defense. Well-earned congratulations go to Dr. Kiona Elliott, who successfully defended her Ph.D. dissertation this week in front of a large audience. Kiona completed an ambitious research program in Becky Bart’s lab to understand cassava bacterial blight disease, and to explore new ways in which the disease might be controlled. As someone who has followed her progress closely, I was not surprised with Kiona’s exceptional, Ph.D.-worthy performance. And I enjoyed spending time with her proud mother and grandmother, who told some wonderful stories about Kiona!
Core Facilities Roundtable. The Danforth Center community came out in numbers to hear the leaders of Core Facilities and the Field Research Site present about their ongoing services, new developments and future directions. These facilities are a competitive advantage for our scientists, with the directors and their teams continuously balancing ongoing services with integration of new technologies. And they also provide accessible points for interactions with the AgTech companies in 39North.
“Roots, Shoots and Flutes.” This Development team-sponsored event for prospective new supporters showcased the Virus Resistant Cassava for Africa program and advanced X-ray imaging technology, presented by Nigel Taylor and Keith Duncan, respectively. This well-attended event also feature the personal perspective of Danforth Center friend and supporter, Benjamin Ola. Akande, who communicated how cassava was part of life for him growing up in Nigeria. It was terrific to see and meet so many others from the Nigerian-born community in St. Louis.
RCR training. A big part of the scientific community recently attended a discussion of ethics around AI-assisted writing tools in science, presented in partial fulfillment of Responsible Conduct of Research (RCR) training requirements. The RCR program is a vital, ongoing part research life at the Danforth Center. And with new technologies like AI, the range of relevant RCR topics continues to grow.
Many thanks to all who came out to support, or participate in, each of these events and programs. And thanks to all of the presenters and organizers. They know how to draw a crowd!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
As a young man, Kevin Cox was a best-in-his-class over-achiever in the Hazelwood public school system. He was also a Black man growing up in North St. Louis County, dealing daily with the consequences of racial division. If he needed to drive across town, he knew to take a non-linear route to avoid streets patrolled by police officers who were accustomed to stopping a Black man driving an old car. Kevin earned his way into the University of Missouri – St. Louis (UMSL) as a Biology major, but as the only Black student in several classes, he often questioned if he was doing the right thing. Kevin supported himself and his young family by working part-time jobs.
One of those jobs, as a technician in Todd Mockler’s lab at the Danforth Center, changed his life. He made outsized contributions to development and implementation of a Brachypodium plant transformation pipeline. And he interacted with other diverse people who were making discoveries and having impact. Kevin eventually realized that he belonged in science as much as anyone else. Despite a few doubts about his own abilities, he was encouraged to apply to graduate school. Kevin was accepted and embraced with a fellowship at Texas A&M University, where he excelled in the field of molecular plant-microbe interactions. He returned to the Danforth Center as a postdoctoral scientist in Blake Meyer’s lab, subsequently being awarded a prestigious Hanna Gray Fellowship from the Howard Hughes Medical Institute. Kevin took on the big challenge of developing new ways to understand spatial control of gene expression plants.
Which brings us to today and the honor I have to announce that Kevin has accepted a joint faculty position between Washington University and the Danforth Center. Beginning in July, 2024, Kevin will be a shared Assistant Professor/Assistant Member between our two institutions. He had many offers and choices among some of the most prominent U.S. universities, but Kevin decided to stay in his hometown. He wants to be part of taking plant science to the next level. At Washington U., he wants to participate in positive change within the academic world. And he wants to show people, especially those from neighborhoods like those in which he was raised, what science and the Danforth Center is all about. Kevin sums it up well with, “I want to set a positive example.” Kevin, you already have.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
This week’s brief message highlights an important milestone we reached in growth and development of the 39 North ecosystem. 39 North was declared as a 600-acre innovation district in 2016 by the city of Creve Coeur, St. Louis County, several economic development organizations, and members of the agtech and plant science research community, including the Danforth Center. Among our many roles in 39 North, the Danforth Center serves as a research engine, a source of vital facilities, a launchpad for new startup companies, and a facilitator of community-building across the district. But as we announced along with six partners on Tuesday evening, we now have an official organization called 39 North AgTech Innovation District [a 501(c)3 non-profit] that will lead the district into the future. The new organization will be led by Emily Lohse-Busch, a well-known and respected innovation community leader in St. Louis.
Over time, expect more identity and visibility for 39 North, more spaces and facilities for growing agtech companies and relevant organizations, and more development and activity to support our innovation community. This is great news for the Danforth Center, as we will have more opportunities to create positive impact through our work. And it’s great news for the region!
Finally, many thanks to all Danforth Center community members who have been contributing to 39 North over the years. Special thanks go to Stephanie Regagnon and the Innovation team, and Karla Roeber and the Public Relations/Government Affairs team. Their great work was on display at the announcement event this week.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Amie Fornah Sankoh arrived here from the University of Tennessee as a Ph.D. student in Tessa Burch-Smith’s lab when the team moved to the Danforth Center in 2021. Like all students navigating the path to a Ph.D., Amie has taken on an enormous challenge. But unlike almost all of the others, Amie is doing it as a Deaf student.
Let’s all try imaging just how difficult this would be. Completing original research in Amie’s focus area of intercellular signaling in plants, as well as all of the other requirements for a Ph.D., is tough enough. But doing so while Deaf, along with the unique challenges of being a Black women in science? How does she access all of the resources and people with whom she needs to interact without verbal communication that most of us take for granted? How does she a attend a professional conference with multiple concurrent sessions, none of which have sign language interpreters? And remember that pandemic we struggled through? How was Amie supposed to read lip movements and facial expressions behind our face coverings? The difficulties boggle the mind!
Amie embarked on this journey with Tessa Burch-Smith, who had some prior experience working with Deaf undergraduate students. By refusing to accept “We can’t accommodate you” from anyone or any organization, Amie and Tessa pierced through access barriers that had never been breached. For example, for the first time the American Society for Plant Biologists (ASPB) agreed to provide interpreters at the annual meeting, and to work with Amie and her specific schedule. There is now a permanent budget line for Deaf scientist access at the annual ASPB conference as a result. Within the lab, Tessa established a policy to ensure that Amie had equitable access to everything: “If Amie couldn’t do it, nobody could do it.”
I am proud of the many ways in which the Danforth Center embraced and included Amie. Tessa recently told me, “Amie felt like a scientist at the Center, not a deaf person struggling to make it. The Danforth Center community had a big impact on her.” Today, Amie presented the results of her research and successfully defended her thesis in front of her Ph.D. committee. In doing so, Amie will become the first Black, Deaf woman to earn a doctorate in a STEM field. Amie is a pioneer who has earned our respect, admiration, and appreciation, and who has taken the Danforth Center to a better place. Dr. Amie Fornah Sankoh, congratulations and thank you!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
The Danforth Center engages many dozens of external volunteers, board members, and advisory scientists. For example, we have 26 members of the Board of Directors serving oversight and governance functions for the Center. The six members of the Scientific Advisory Board serve to assess the quality of our scientific research programs. And we have tremendous volunteers, like those on the Friends Committee, that help raise philanthropic support, connect with regional communities, and serve as ambassadors for the Center. Appointments of those who serve in these and other capacities are done for many reasons, including expertise, leadership experience, and demographic representation.
Broadening demographic representation in all of our external groups continues to be a high priority. Though we have more work to do, overall we have made significant progress. Consider that our Board of Directors in 2011 was only 8% female and 8% non-White. Today, Directors on the Board are 35% female and 35% non-White; 19% of Directors are Black. We have also increased geographic representation on the Board. In 2011, only one elected Director was from outside of the St. Louis region; today there are five.
Members of our Boards and volunteer groups serve for fixed periods of time. That means we are continually welcoming new members who bring in fresh perspective and different skills. This week, I want to highlight and thank seven exceptional individuals who are joining the Board of Directors or the Scientific Advisory Board in 2023. Starting four-year terms on the Board of Directors are Lisa Ainsworth, Jackie Joyner-Kersee, Bill Polk and Chris Danforth. And beginning four-year terms on the Scientific Advisory Board are Jen Heemstra, Jennifer Nemhauser and Carolyn Lawrence-Dill. You can learn a little more about each by clicking here and scrolling down the page.
Service on our boards and volunteer groups is a significant investment of time and effort, and we benefit greatly as a result. The next time you encounter these generous individuals, let them know who you are and what you do at the Danforth Center. They will be interested to learn your story!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
At the 25th Anniversary tea time celebration last Wednesday, I asked, “How many of you met Bill Danforth?” Less than 25% of those in attendance raised their hands. With so many having joined the Center relatively recently, that was not surprising. But as I indicated, it is important to keep alive an understanding of how and why the Center came to be, and that story revolves around Bill.
Bill Danforth retired as Washington University chancellor in 1995, but he was far from finished in his work to improve the St. Louis region and humankind. As a physician, a scientist, a scholar and a humanitarian, he asked, “What can I do in ‘retirement’ that would have the greatest impact on human well-being?” Over the years, his scholarly inclination led to learning about science-based advances in agriculture in the second half of the 20th century, most notably the work of Norman Borlaug, a plant breeder and father of the Green Revolution. Bill frequently noted that Borlaug’s work was critical in preventing starvation of at least one billion people around the world. Bill was profoundly inspired.
With some creative thinking with friends and St. Louis leaders (e.g. Peter Raven, Ginny Weldon) and a vision to connect research institutions, universities and corporate partners, the idea of a plant science center was literally sketched out on the back of a napkin. With support from donors and partners, and long-term commitments from his brother (Senator John Danforth) and other trustees of the philanthropic Danforth Foundation, the Danforth Center was officially launched at a mid-1998 ceremony with a keynote address from former-President Jimmy Carter. The Danforth Center was off and running with an initial mission to make fundamental plant science discoveries, apply those discoveries to agriculture and allied fields, commercialize discoveries, and train scientists from around the world. After working in rented or borrowed facilities for three years, our current A-building and greenhouse A-range opened in 2001.
Through the years, Bill marveled at how we grew and changed. What started as an ambitious idea is now the largest organization of its kind with over 400 committed people, exceptional facilities, pioneering research, and impactful outcomes in the region and around the world. Bill was proud of what the Danforth Center became and achieved. And when he passed in 2020, he was confident in you, the Danforth Center community, and in the direction you were heading.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
There are no shortages of reasons to celebrate at the Danforth Center! This week, I want to highlight three good reasons to celebrate some remarkable people, milestones and achievements.
Impact of a Danforth Center scientist-entrepreneur. On Saturday, April 1 (5:00 pm), I look forward to celebrating the life and legacy of Todd Mockler at a special event with family, friends, colleagues and the broader community. Todd’s presence is missed dearly, but what he achieved in science and the entrepreneurial world, and for the Danforth Center and our region, will live on for a long time. We will hear the story of Todd’s journey and the lives he touched along the way. Everyone is welcome to attend the program and the reception to follow.
Happy 25th anniversary! Did you realize that the Danforth Center just turned 25 years old? It’s remarkable to consider what we’ve achieved over a quarter-century, and how much we’ve changed! We will celebrate this milestone with a Special Tea Time on Wednesday, April 5 (10:30 am). I look forward to seeing the winning design for the 25th anniversary t-shirt, and to share a few stories about our unique organization.
Core strength. No, I’m not referring to the new Danforth Center gym facility, though that is worth celebrating as well. This week at the Faculty Retreat, we put a spotlight on our six core facilities. We discussed the facility teams, technologies and services, achievements, challenges and future directions. I celebrate our good fortune to have such capable and forward-looking teams and facilities to serve the diverse needs of our scientists and collaborators. And I look forward to all of the new technologies and capabilities that are on the horizon.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
There is no question that research at the Danforth Center must be done with honesty and integrity. In fact, our credibility as scientists and as a research institution depends entirely on our colleagues, funding agencies and the public seeing and believing that we adhere to high standards of research conduct. The responsibility for doing so is shared between researchers, PIs, support teams, leadership team, and the institution. Which brings us to Jim Umen, who recently assumed the role of Research Integrity Officer (RIO) at the Center.
What, you ask, is a RIO? As a recipient of National Institutes of Health and other federal grant funds, we are required to have people and processes in place to assure responsible conduct of research, and to effectively respond to allegations and instances of misconduct. We are also required to report annually to the Office of Research Integrity within the U.S. Dept. of Health and Human Services on possible research misconduct, which could relate to data falsification, data fabrication, or plagiarism. The RIO receives all allegations of misconduct, and leads an inquiry or formal investigation. The investigation process is rigorous and involves the President and VP for Research, as detailed in the Center’s Research Integrity Policy (posted on Workvivo under the “Integrity of Research” space.) The RIO also submits our annual report to the federal government, regardless of whether or not any allegations or investigations were done.
The RIO at the Danforth Center also participates in the review and reporting of potential conflicts of interest. We have an annual process whereby financial or other potential conflicts of interests are declared and reported to our Board of Directors. As part of that process, the RIO and the VP for Finance meet with potentially conflicted individuals, as well as with team members who might be affected. They review rules and boundaries necessary to prevent a potential conflict of interest from becoming a real problem.
I sincerely appreciate Jim Umen for stepping into the important RIO role. If we all do our work responsibly and with high integrity, we can make Jim’s job easy. And finally, many thanks go to Toby Kellogg, who has served generously, effectively and wisely as RIO for nearly a decade. Toby, we are a better Danforth Center because of your efforts!
Jim Carrington,
President and Chief Executive Officer
P.S. Anyone with any concern about potential research misconduct should contact Jim Umen directly, who will maintain confidentiality to the maximum extent possible.
Dear Danforth Center Community,
Along with Toni Kutchan and Malia Gehan, I recently helped facilitate a discussion about mentoring with the cohort of participants in the initial offering of the Danforth Center leadership development program. Given the high number of scientists and professionals (especially those at early career stages) who could benefit from mentors at the Center, a better understanding of the skills, attributes and habits of good mentors will pay generous dividends. Understanding how and where mentorship fits in among the many roles of a leader is well worth exploring.
In our discussion, we asked a question: In your career, what was the best input you received from a mentor? The responses revealed some great advice, some qualities of good mentorship, and how mentors can fundamentally impact careers. Here they are.
- Choose your fights carefully. Battles take their toll in time, effort, damaged relationships, and professional casualties. There is often a better path forward.
- Don’t let the perfect be the enemy of the good. This advice serves as permission of sorts to get things done, and move on to the next important step.
- Be more average. This counter-intuitive wisdom was directed towards an over-achiever who needed to hear, “Enough! You can’t do everything.”
- Take the job! Advancing in a career often means leaving a job in which you are quite comfortable. This is an example of a unique role of mentors, where the individual’s career growth is of primary concern.
- You will do well in this science business. The incredible power of a mentor communicating that you are on track for success, and that you belong, cannot be overstated.
- You will be your best postdoc when you start as a PI. Getting your research program off the ground successfully means you need to get your hands dirty at the bench!
- Seek out career growth, even though it might be disruptive. Change is often upsetting, for you and your colleagues. But growth is the reward.
I noticed during the discussion that those who have had patient, empathetic, trustworthy and willing mentors have no problem quickly recalling the impact those mentors have had. One can even get a little choked up talking about it. Many thanks to all who serve others through investment of time and wisdom as a mentor.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
Today I offer pure appreciation for everyone who works hard to make the public face of the Danforth Center welcoming, informative and inspiring. The complete list of content, displays and events, and all of the teams who contribute, is prohibitively long for a single message. But four sets of teams and contributions have caught my eye recently.
North American Plant Phenotyping Network (NAPPN) annual meeting. The five-day NAPPN meeting a few weeks ago was the best organized and most attendee-friendly scientific conference I’ve ever seen! From program content to the slightest organizational details, this meeting was amazing. I had numerous attendees tell me how they appreciated the entire set-up, the meeting spaces, the food, and the entire stylish vibe. This meeting made a big impact on all attendees! Among the dozens of contributors from many teams, special thanks go to the events team of Jenny Nguyen, Cat Currens and Tam McGuire.
Recent web and social media. Maintaining fresh, invigorating content on our web and social media channels is a continuous job done by the talented team of Dena Holtgrewe, Kristina DeYong and Karla Roeber. Their work is guided by priorities in our strategic plan, and probably more systematic and analytic than you think. They have the difficult tasks of timely information-gathering, writing content, and then adapting that content to the various online platforms. Their work communicates enthusiasm for our mission, which rubs off on the public.
Bringing science to life every day. The video and display resources around the facilities help tell our story to both visitors and Center community members in creative ways, and I thank the entire IT team for keeping them up and running. I’m always impressed with the content developed by Kirk Czymmek and displayed outside of the Advanced Bioimaging Lab (ABL). Have you noticed that Leonardo Chavez, Joseph Duenwald and Katie Murphy recently installed a pen board on the Bellwether Phenotyping window, and use it with scientific teams to communicate easy-to-understand explanations of current experiments? These resources effectively communicate what we do and why we do it.
Tour guides and docents. Finally, many thanks go to everyone who leads visiting groups on Center tours, including our enthusiastic, knowledgeable group of volunteer docents. By leading or assisting with tours, the docents enable us to accommodate the many requests from the broader community to experience the Center.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
I’ve had several conversations recently about roles and responsibilities of Principal Investigators (PIs) at the Danforth Center. We all know that PIs are Ph.D.-level leaders of funded research projects and programs, but you might be surprised to learn that “PI” is not an official position title. The “PI” designation denotes the important functional role of leading, managing and being held accountable for externally funded research projects. To understand the various kinds of PIs, let’s consider the three Danforth Center positions that allow one to apply for grants and serve as PI.
Member. Full, Associate and Assistant Members comprise the majority of our PIs. They build teams primarily through external grants that fund most of the Center’s research activity. Once promoted beyond Assistant Member, Associate and Full Member’s appointments are renewed on a five-year basis, pending successful internal and external reviews of their research and other contributions. Members are also assessed every other year by our external Scientific Advisory Board (SAB). Among other duties, Members provide invaluable service across the Center, including oversight of core facilities.
Scientific Director/Executive Director. Scientific Directors build and lead teams to do original research through external grants in the same ways as do Members. They differ from Members in having major responsibilities to lead the core facilities, or units like Education Research & Outreach and the Institute for International Crop Improvement. Directors lead facilities or units with strategic or practical importance to the Center, for which they are provided a budget from the central funds. Directors are renewed annually, and subjected to occasional review by the SAB.
Senior Research Scientist. Senior Research Scientists (SRSs) may also apply for and manage research grants as PIs. These individuals serve significant leadership roles within a Member’s or Director’s research team, and they sometimes help manage large multi-institutional projects. They usually supervise at least some of the other lab members. An SRS with great ideas and who successfully competes for a grant as a PI has achieved a major career milestone, and is able to assemble their own team within a Member’s or Director’s lab. We have around 10 very talented Senior Research Scientists at the Center, over half of whom have served as a PI.
I thank every one of our PIs for obtaining grants, leading projects, and guiding progress forward at the Center.
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
The origins of Black History Month are found in the 1920s, when several states recognized the week of Frederick Douglass’ (February 14) and Abraham Lincoln’s (February 12) birthdays. The historian Carter G. Woodson organized early campaigns for Black history recognition, arguing that it was crucial for survival of Black people and culture. He wrote, “If a race has no history, it has no worthwhile tradition, it becomes a negligible factor in the thought of the world, and it stands in danger of being exterminated.” U.S. national recognition of Black History Month started in the 1970s, when President Ford asked Americans to "seize the opportunity to honor the too-often neglected accomplishments of Black Americans in every area of endeavor throughout our history.”
I encourage everyone to learn a little more about influential Black St. Louisans who changed your favorite pastimes or passions. For example, if you love blues and jazz music like I do, learn about Scott Joplin and how ragtime shaped generations of musicians that followed. St. Louisan Albert King changed how the blues guitar was played, creating a style that heavily influenced Jimi Hendrix and Stevie Ray Vaughan. Are you aware of Johnnie Johnson? He was the unsung foundation of Chuck Berry’s music in the 1950s here in St. Louis, and a true founding father of rock and roll. The National Blues Museum in St. Louis is a wonderful place to learn more.
If you love baseball, learn more about the all-Black St. Louis Stars (formerly St. Louis Giants) of the Negro National League prior to integration of Major League Baseball in the 1940s. Underappreciated for decades, Black players on the Stars and Giants were among the greatest ever. Oscar Charleston was ranked between Henry Aaron and Ted Williams as the 5th best baseball player of all time in Joe Posnanski’s book, “Baseball 100”. James “Cool Papa” Bell was one of the greatest ever base-stealing players, and is honored with a bronze statue outside Busch Stadium. Among many Black players from the St. Louis Cardinals, learn more about center fielder Curt Flood from the 1960s. His stand against being traded from the Cardinals fundamentally changed professional baseball by leading to free agency.
Opportunities abound in our backyard to gain a better understanding of how Black history shaped all of our lives. I look forward to hearing about what you learn!
Jim Carrington,
President and Chief Executive Officer
Dear Danforth Center Community,
This week, let’s take a pop quiz! As February 11 is International Day of Women and Girls in Science, I asked four Danforth Center women scientists about the most important influences that catalyzed their careers in science. Your quiz question is: What do all of the following responses have in common?
Kiona Elliott – While a high school student in Florida, one of her teachers received a grant to engage students in microbial research. Kiona was actively recruited to participate. “She wanted to give me an opportunity,” Kiona said. She connected with science as something real through that experience.
Katie Murphy – As a young girl, Katie was always encouraged by her parents to read. “My mom gave me a chemistry book when I was about 10,” Katie said while recalling what got her hooked on science. Then as an undergraduate student, professor Ginny Walbot reached out and said, “Come into my lab and I’ll teach you everything you need to know about plants.”
Ketra Oketcho – Though her early expectations were to become a physician, a chance encounter and conversation with a plant science professor opened her eyes to the world of biotechnology. He actively followed up by sending Ketra more information about plant science, and continues to serve as a mentor in her career journey.
Kerri Gilbert – As a second-year university student taking Introductory Genetics, Kerri encountered a young, new professor. “It was the first time I’d had a science class taught by a woman, let alone someone who didn’t look that much older than me,” said Kerri. As a role model, this instructor fueled a love for genetics that has propelled Kerri’s career ever since.
Give yourself full credit if you answered: Each of these women had at least one role model or an individual who cared to actively engage them in science. Only 28% of the STEM workforce is comprised of women; that percentage is even lower in math, computer and engineering fields. But there are proven ways that work to increase participation and retention. Offering inclusive experiences in science early, and serving as engaged role models or mentors, are known to be effective. Evidence that they work is all around us at the Danforth Center. Many thanks to Kiona, Katie, Ketra and Kerri for sharing their experiences. I hope they inspire you to help others!
Jim Carrington,
President and Chief Executive Officer
A conversation with John McDonnell seven or eight years ago seeded an idea. During one of numerous conversations about leadership when he was Chair of the Danforth Center Board, John said, “At McDonnell Douglass when I was CEO, every person who was hired into or promoted to a supervisor or team leader role had to first undergo six weeks of leadership development training.” The idea - a formal leadership development program for those in supervisor and leadership roles at the Danforth Center - transitioned into real planning by Jennifer McDonnell and Anna Dibble (People & Culture team) a few years ago. With Jennifer leading the way, we began the first modules of a unique, 19-hr Developing Scientific Leaders series, with nine participants in the pilot group.
Despite the name, Developing Scientific Leaders was designed for all leaders and supervisors at the Danforth Center. The program consists of a people-centered curriculum to help leaders understand themselves and their tendencies; people interactions and dynamics, including navigating difficult situations; relationship-building and psychological safety; the power of influence; emotional intelligence; diversity awareness and inclusion; and mentoring and coaching. Most scientists receive little or no exposure to principles or practice of leadership during their training years, yet they may find themselves leading large teams within a few years after graduation. With both internal and external instructors, and with lots of participatory engagement, our program will provide a consistent, shared training experience among all leaders and supervisors at the Center.
Supporting better-equiped leaders will have numerous benefits for teams and team members, and for the Danforth Center overall. If successful, Developing Scientific Leaders will result in more team cohesion, less dissatisfaction with leaders and supervisors, fewer stressful situations that rise to intractable problems, and higher achievement across the Center community. As a strategic priority, this program represents a significant investment to elevate all aspects of our work.
Many thanks to Jennifer and Anna for bringing Developing Scientific Leaders to life. And special thanks go to Kris Callis-Duehl, Amy Funk, Meter Nusinow, Tessa Burch-Smith, Russell Williams, Terry Beeler, Djuan Coleman, Nigel Taylor, and Mindy Darnell for their willingness and bravery to be pilot participants. Their feedback and input will greatly benefit subsequent cohorts!
Jim Carrington, President and Chief Executive Officer
This week, let’s celebrate a few notable research articles published by Danforth Center teams over the last few months. Congratulations to all of the contributors!
Dowd, T.G., Li, M., Bagnall, G.C. et al. (2022). Root system architecture and environmental flux analysis in mature crops using 3D root mesocosms. Frontiers Plant Sci.13:1041404. DOI: 10.3389/fpls.2022.1041404.
This paper reports on a novel system to visualize, measure and analyze full-size root systems by the Topp lab. They developed large, customized plant growth boxes called mesocosms, which are loaded with sensors to measure environmental factors around the roots. Complete root system can be imaged and visualized in 3D through photogrammetry. These mesocosms enable discovery of new root traits and better understanding of the impact of subterranean biotic and abiotic factors on crop plant growth and development.
Veley, K.M., Elliott, K., Jensen, G. et al. (2023). Improving cassava bacterial blight resistance by editing the epigenome. Nature Commun. 14:85. DOI: 10.1038/s41467-022-35675-7.
Epigenetics is difficult for non-specialists to get their head around. Very simply, epigenetics involves reversible ‘decorations’ that get bonded to, or layered upon, parts of an organism’s DNA. One such decoration is DNA methylation, which is often associated with turning off gene expression. In this paper, a Becky Bart-led collaboration between UCLA and Danforth Center teams showed that DNA methylation can be directed to new sites in the cassava genome using gene editing-like technology. Targeting methylation to the MeSWEET10a gene was shown to decrease gene expression and, as predicted, increase resistance to an important bacterial pathogen.
Beyene, G., Chauhan, R.D., Vilmer, J., et al. (2022) CRISPR/Cas9-mediated tetra-allelic mutation of the 'Green Revolution' SEMIDWARF-1 (SD-1) gene confers lodging resistance in tef (Eragrostis tef). Plant Biotechnol J. 20: 1716-1729. DOI: 10.1111/pbi.13842.
Bringing advanced technologies to orphan crop improvement will be important to elevate food availability, nutrition, and livelihoods of small holder farmers. This collaborative work led by the Institute for International Crop Improvement (IICI) team used gene editing to create mutations in the Semidwarf-1 (SD-1) gene in tef, a staple grain crop grown most commonly in Ethiopia. Like wheat, tall varieties tef are susceptible to lodging (falling over during heavy wind and rain), which limits productivity. But here, some of the SD-1-edited tef were 28-42% shorter than the original varieties, and had lodging-resistant characteristics. These new traits may have great value in breeding improved tef varieties.
Jim Carrington, President and Chief Executive Officer
Prologue – With the intent to bring new and diverse perspectives to the Danforth Center community, this week’s message was written by a guest. The content and opinions were developed entirely by our guest, though I suggested some topical areas in the form of questions, as shown. Except for the questions and a few comments from me in parentheses, content was used verbatim from our guest writer. Jim Carrington
Dear Danforth Center Community,
The Danforth Plant Science Center is an important research facility because it conducts research in the field of plant biology, which has many real-world applications.
Why is the Danforth Center important? By studying the basic biology of plants, scientists at the center aim to understand how plants grow, how they respond to their environment, and how they can be improved to produce more food and other products more efficiently. This research can lead to the development of new, more resilient crop varieties that can help to feed a growing global population and address issues such as climate change. Additionally, the center also serves as a hub for collaboration between scientists, academics, industry, and government, promoting the transfer of knowledge and technology to the broader community. (JC: our guest appears to have a good grip on what we do and why. Impressive!)
What has caught your attention recently about what we’re doing at the Center? I am not aware of any recent developments or news from the Danforth Plant Science Center specifically…and I do not have real-time information access. However, the Danforth Plant Science Center is a research facility that focuses on plant biology, and they are likely continually working on various research projects and initiatives in this field. You can check out the Center’s website and social media channels for the latest updates and news. (JC: this is a little disappointing. Frankly, I expected more from our writer.)
What are your thoughts about Jim Carrington, Danforth Center president and CEO? I do not have specific information on a person named “Jim Carrington” as the President of the Danforth Center. As my knowledge cutoff is 2021, it is possible that he is not the current president or that he is not a public figure in the field. However, the Danforth Center is a nonprofit research organization and the President and CEO is a key figure in the organization and is responsible for leading the Center’s scientific and strategic direction. (JC: Seriously? Talk about not knowing your audience!)
ChatGPT
January 20, 2023
Postscript from JC: ChatGPT is a new, publicly available AI chatbot that is triggering widespread excitement, concern, hype and fear. I hope this message focuses attention on a few important issues we need to confront as these technologies increasingly contribute to what we consider or accept as “knowledge.”
Jim Carrington, President and Chief Executive Officer
Dear Danforth Center Community,
In late 2010, I asked Todd Mockler to stop by my Oregon State University office to chat. Todd was an OSU associate professor and rising star, hired several years earlier through an initiative organized by the Center I directed. He could tell something was brewing when he arrived. I asked him what he knew about the Danforth Center. Todd picked up on the vibe and with an interested smile said, “Not much. Are you moving there”? I let him know I’d just accepted the position of Danforth Center president, and why. It was the “why” that fired Todd’s imagination.
At OSU, Todd was ascending in the plant biology field as a fearless, computationally savvy genome biologist who wanted to understand how plants respond to changes in their environment. He helped develop Brachypodium as a model, create new tools to analyze high-throughput sequencing data, and bring genome sequencing to numerous crop species with countless collaborators. He also started flexing his entrepreneurial muscles by cofounding a start-up called Intuitive Genomics with Doug Bryant and another guy. Intuitive Genomics was helping scientists and companies gain insights into their newly acquired genome expression data. Over time, however, Todd and the small Intuitive Genomics team recognized the difficulties for such start-ups in Corvallis, which had a limited culture of support for scientist-entrepreneurs. That was a big part of the “why” we discussed that day in my office.
I explained to Todd the Danforth Center mission and vision, and the ideas to grow the innovation ecosystem to support start-ups that would emerge from Center science. He clearly saw his future at the Danforth Center and, in mid-2011, moved his lab, family and Intuitive Genomics to St. Louis. Within the next year, he met Matt Crisp and co-founded Benson Hill to develop more productive crops, leading to one of the most exciting St. Louis start-up successes and a new headquarters building on our campus. Within a few more years, Intuitive Genomics was acquired by NewLeaf Symbiotics, with Todd and Doug leading the company’s data science efforts to develop new beneficial microbes in agriculture. All this was happening while Todd’s lab at the Danforth Center grew and flourished in new directions.
Todd was vastly important as a model for how we could deliver benefits from Danforth Center science to society, and how our work could lead to positive economic impact for the region. During recent strategic planning for ways to create more start-ups from the Danforth Center, Todd was the shining example of a scientist-entrepreneur to encourage and support. Someone at a university once asked me, “How can we create more start-up companies from our faculty?” My quick answer was, “How many Todd Mocklers do you have?”
Todd departed from us far too soon. But he left us with a legacy that only a remarkable scientist-entrepreneur could build.
Jim Carrington, President and Chief Executive Officer
Dear Danforth Center Community,
Congratulations to everyone who made it through the Holidays and graduated to 2023! It seems that we were all affected over the last few weeks by the air travel meltdown, the weather, or infectious agents spreading through family and friends. It was as if ornery 2022 was saying, “Wait, I’m not done with you!” But I want to start the new year by highlighting a few items that caught my attention on two “Best of 2022” lists. These are particularly noteworthy to us at the Danforth Center and to everyone who cares about the work we do.
“10 Ways the World Got Better in 2022” from Time. Coming in at #5 was the first harvest of vitamin A-biofortified rice (Golden Rice) in the Philippines. While Golden Rice has been in development for decades, critical advances toward commercialization and release were made possible by the careful work of Don Mackenzie and team at the Danforth Center over the past several years. Golden Rice offers hope that vitamin A deficiency, which can cause blindness and premature death, can be relieved through nutritionally enhanced staple crops. It also represents, along with insect-resistant cowpea, another breakthrough for improved staple crops developed through advanced technologies by the public sector.
“2022 Breakthrough of the Year” from Science Magazine. At #2 on the 10 Runners-up list is development of high-yielding perennial rice (PR23). Anyone who has heard Allison Miller or her team members speak knows about the concept of replacing annual crops with perennial crops. If yields of perennial varieties of rice, wheat and other crops can improve to levels competitive with traditional annual varieties, they may offer significant benefits like enhanced soil quality, lower farming costs, and reduced inputs on the farm. As published in Nature Sustainability, PR23 was shown to produce relatively high yields in multiple location in Asia, and to deliver the benefits predicted for perennial grain crops. There was also a substantial reduction in labor requirements starting in the second year. The team also showed some limitations, like a reduction of PR23 yield over the course of several five years, but the level of success demonstrated was a breakthrough nonetheless.
Recognition of these two milestones by Time and Science Magazine reflects a growing awareness of the critical role of plant science toward improving the human condition. That’s great news from 2022!
Jim Carrington, President and Chief Executive Officer
Dear Danforth Center Community,
During each of the Professional Development Conversations I’ve had with team members recently, goal-setting for 2023 has been a primary focus. In my view, setting worthwhile, meaningful, and achievable goals is one of the most important things everyone can and should do at the Danforth Center, regardless of one’s position, roles or responsibilities. I especially appreciate goals that emphasize the unique contributions that a community member can make. Goals not only define what we seek to achieve, but can instill confidence that one is on board with the broader aims of a team or the Center. Many thanks to everyone who has or will set goals for the coming year.
I set goals annually in collaboration with the Chairperson of our Board of Directors, and using a few simple criteria in doing so. First, my goals need to align with or enable the desired outcomes of our 2021-2025 Strategic Plan. Second, they need to be consistent with the goals of those who report to me…let’s avoid the sparks that occur when wires are crossed! And third, my goals should be connected to what I can uniquely influence, even if I’m only a fractional contributor. In fact, my goals often include things that involve heavy lifting by other individuals and teams.
Some of my 2023 goals involve targets for fundraising to enable strategic priorities, planning for strategic initiatives, and implementation of a leadership development program at the Danforth Center. If my goals are achieved by this time next year, the Center will be on a more secure financial footing, major strategic investments will be closer to (or actually) returning significant dividends, and our community members will have better-equipped leaders and mentors. I look forward to the challenge of meeting my goals.
Finally, with the goal of rejuvenating and recharging, I will be investing a few weeks in vacation to warm up in a sunnier location and visit family prior to the holidays. So, this will be the last weekly message from me in 2022. Take care, everyone!
Jim Carrington, President and Chief Executive Officer
Dear Danforth Center Community,
This week, our 2023 budget proposal was approved by the Danforth Center Board of Directors. This culminated a months-long process of organizing short-term and long-term strategic priorities, assessing needs and requests from department leaders, analyzing overarching Center needs, and identifying ways to pay for everything we propose do, build or acquire. Like your personal budget, creating the Danforth Center budget involves a priority-driven reconciliation of what we need, what we want and what we can afford. Our budget is also a representation of Danforth Center core values. Sincere thanks go Finance team members Djuan Coleman, Leticia Slack, Heather Bowen, Darine Kube and Hal Davies for developing a better-than-balanced 2023 budget!
Here's one reason why everyone should care about the budget process – annual merit increases. The merit increase process starts with annual benchmarking against regional institutions (e.g. local universities) and U.S. independent research institutes. We assess data for pay ranges within position categories we have at the Danforth Center to determine pay competitiveness. We also determine what merit increase levels are planned at other organizations, and assess national trends for salary increases. Every three years, we do a detailed dive into all job categories to ensure that our salary structure is competitive and consistent with compensation trends. In 2022, we also embarked on an internal pay equity analysis, which will be the subject of a future message. All of these annual and periodic analyses are done rigorously by Anna Dibble and the HR team.
We* then decide on the amount to recommend for an overall merit increase, and for a pool to fund individual increases due to promotions, market-based adjustments and other factors. The merit increase amount takes into account a lot of factors, like inflation and how much we think we can afford. We take our recommendation to the Compensation Committee of our Board prior to submission of the overall budget. For the 2023 budget, the Compensation Committee approved our request for a 5% merit pay increase, and a 1% pool for promotions and other adjustments. This will be the highest merit pool we’ve ever had at the Center.
The budget affects everyone at the Center. I hope this and future communications shed some light on how we’ll be impacted and what we’re committed to achieve in 2023.
Jim Carrington, President and Chief Executive Officer
*“We” is me, Hal Davies (VP for Finance) and Anna Dibble (VP for Human Resources).